6O2N
 
 | |
5HYI
 
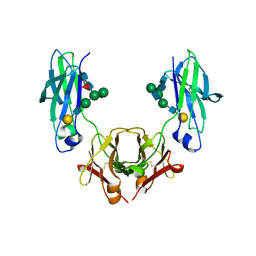 | | Glycosylated, disulfide-linked Hole-Hole Fc fragment | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural differences between glycosylated, disulfide-linked heterodimeric Knob-into-Hole Fc fragment and its homodimeric Knob-Knob and Hole-Hole side products.
Protein Eng. Des. Sel., 30, 2017
|
|
7UWL
 
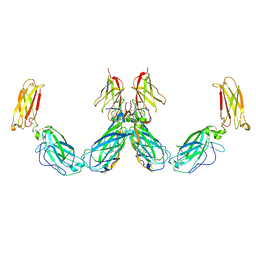 | | Structure of the IL-25-IL-17RB-IL-17RA ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWM
 
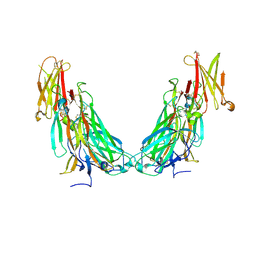 | | Structure of the IL-17A-IL-17RA binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWK
 
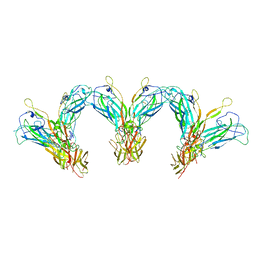 | | Structure of the higher-order IL-25-IL-17RB complex | | Descriptor: | Interleukin-17 receptor B, Interleukin-25 | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWJ
 
 | | Structure of the homodimeric IL-25-IL-17RB binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor B, Interleukin-25 | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
6NWW
 
 | |
6C63
 
 | | Crystal Structure of the Mango-II Fluorescent Aptamer Bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, POTASSIUM ION, RNA (32-MER), ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-01-17 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.900028 Å) | | Cite: | Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Biochemistry, 57, 2018
|
|
5XP0
 
 | |
7UWN
 
 | | Structure of the IL-17A-IL-17RA-IL-17RC ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7DMU
 
 | |
8JMR
 
 | | Crystal structure of hinokiresinol synthase in complex with 1,7-bis(4-hydroxyphenyl)hepta-1,6-dien-3-one | | Descriptor: | 1,7-bis(4-hydroxyphenyl)hepta-1,6-dien-3-one, Hinokiresinol synthase alpha subunit, Hinokiresinol synthase beta subunit, ... | | Authors: | Ding, Y, Ushimaru, R, Mori, T, Abe, I. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insights into the C-C Bond-Forming Rearrangement Reaction Catalyzed by Heterodimeric Hinokiresinol Synthase.
J.Am.Chem.Soc., 145, 2023
|
|
8JMQ
 
 | | Crystal structure of hinokiresinol synthase | | Descriptor: | Hinokiresinol synthase alpha subunit, Hinokiresinol synthase beta subunit | | Authors: | Ding, Y, Ushimaru, R, Mori, T, Abe, I. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into the C-C Bond-Forming Rearrangement Reaction Catalyzed by Heterodimeric Hinokiresinol Synthase.
J.Am.Chem.Soc., 145, 2023
|
|
6CQ6
 
 | | K2P2.1(TREK-1) apo structure | | Descriptor: | CADMIUM ION, DECANE, POTASSIUM ION, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2018-03-14 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | K2P2.1 (TREK-1)-activator complexes reveal a cryptic selectivity filter binding site.
Nature, 547, 2017
|
|
2B00
 
 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Glycocholate | | Descriptor: | CALCIUM ION, GLYCOCHOLIC ACID, Phospholipase A2, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
2B04
 
 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Glycochenodeoxycholate | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCOCHENODEOXYCHOLIC ACID, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
7CY1
 
 | | Crystal Structure of MglC from Myxococcus xanthus | | Descriptor: | Mutual gliding motility protein C, SODIUM ION | | Authors: | Thakur, K.G, Kapoor, S, Kodesia, A. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural characterization of Myxococcus xanthus MglC, a component of the polarity control system, and its interactions with its paralog MglB.
J.Biol.Chem., 2021
|
|
5LSB
 
 | | Crystal structure of yeast Hsh49p in complex with Cus1p binding domain. | | Descriptor: | Cold sensitive U2 snRNA suppressor 1, Protein HSH49 | | Authors: | van Roon, A.M, Obayashi, E, Sposito, B, Oubridge, C, Nagai, K. | | Deposit date: | 2016-08-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain.
RNA, 23, 2017
|
|
6NG0
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with sunitinib in the inactive state. | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Johnson, E, McTigue, M, Cronin, C.N. | | Deposit date: | 2018-12-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Multiple conformational states of the HPK1 kinase domain in complex with sunitinib reveal the structural changes accompanying HPK1 trans-regulation.
J.Biol.Chem., 294, 2019
|
|
6CQ9
 
 | | K2P2.1(TREK-1):ML402 complex | | Descriptor: | CADMIUM ION, HEXADECANE, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2018-03-14 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | K2P2.1 (TREK-1)-activator complexes reveal a cryptic selectivity filter binding site.
Nature, 547, 2017
|
|
8J54
 
 | | Crystal structure of RXR/DR2 complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*CP*CP*TP*AP*CP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*GP*GP*TP*CP*AP*GP*TP*AP*GP*GP*TP*CP*AP*TP*G)-3'), Retinoic acid receptor RXR, ... | | Authors: | Chen, Y, Jiang, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural characterization of the DNA binding mechanism of retinoic acid-related orphan receptor gamma.
Structure, 32, 2024
|
|
5JW9
 
 | |
5JZ7
 
 | | NGF IN COMPLEX WITH MEDI578 scFv | | Descriptor: | Beta-nerve growth factor, MEDI578 scFv, heavy chain, ... | | Authors: | Olsson, L.-L, Aagaard, A. | | Deposit date: | 2016-05-16 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Engineering the surface properties of a human monoclonal antibody prevents self-association and rapid clearance in vivo.
Sci Rep, 6, 2016
|
|
6ZIQ
 
 | | bovine ATP synthase stator domain, state 1 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TNG
 
 | | Structure of FANCD2 in complex with FANCI | | Descriptor: | Fanconi anemia complementation group I, Uncharacterized protein | | Authors: | Alcon, P, Shakeel, S, Passmore, L.A. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
