6T2H
 
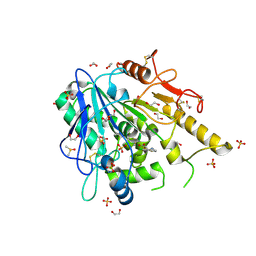 | | Furano[2,3-d]prymidine amides as Notum inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-[[(4~{S})-5-chloranyl-6-methyl-1,2,3,4-tetrahydrothieno[2,3-d]pyrimidin-4-yl]sulfanyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Scaffold-hopping identifies furano[2,3-d]pyrimidine amides as potent Notum inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4F9D
 
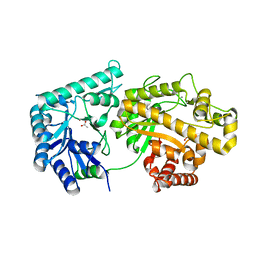 | | Structure of Escherichia coli PgaB 42-655 in complex with nickel | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Little, D.J, Poloczek, J, Whitney, J.C, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure and Metal Dependent Activity of Escherichia coli PgaB Provides Insight into the Partial De-N-acetylation of Poly-b-1,6-N-acetyl-D-glucosamine
J.Biol.Chem., 287, 2012
|
|
4F9J
 
 | | Structure of Escherichia coli PgaB 42-655 in complex with iron | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Little, D.J, Poloczek, J, Whitney, J.C, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | The Structure and Metal Dependent Activity of Escherichia coli PgaB Provides Insight into the Partial De-N-acetylation of Poly-b-1,6-N-acetyl-D-glucosamine
J.Biol.Chem., 287, 2012
|
|
3W2R
 
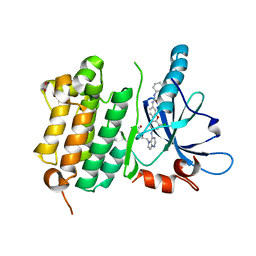 | | EGFR Kinase domain T790M/L858R mutant with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 1-{3-[2-chloro-4-({5-[2-(2-hydroxyethoxy)ethyl]-5H-pyrrolo[3,2-d]pyrimidin-4-yl}amino)phenoxy]phenyl}-3-cyclohexylurea, Epidermal growth factor receptor | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
6KNB
 
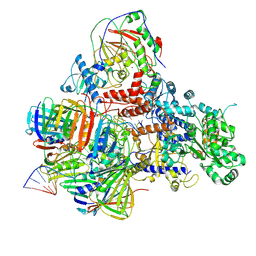 | | PolD-PCNA-DNA (form A) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
4GU9
 
 | |
3W2S
 
 | | EGFR kinase domain with compound4 | | Descriptor: | 1-{3-[2-chloro-4-({5-[2-(2-hydroxyethoxy)ethyl]-5H-pyrrolo[3,2-d]pyrimidin-4-yl}amino)phenoxy]phenyl}-3-cyclohexylurea, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
1I1O
 
 | |
2K71
 
 | | Structure and dynamics of a DNA GNRA hairpin solved vy high-sensitivity NMR with two independent converging methods, simulated annealing (DYANA) and mesoscopic molecular modelling (BCE/AMBER) | | Descriptor: | 5'-D(*DGP*DCP*DGP*DAP*DAP*DAP*DGP*DC)-3' | | Authors: | Santini, G.P.H, Cognet, J.A.H, Xu, D, Singarapu, K.K, Herve du Penhoat, C.L.M. | | Deposit date: | 2008-07-29 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nucleic acid folding determined by mesoscale modeling and NMR spectroscopy: solution structure of d(GCGAAAGC).
J.Phys.Chem.B, 113, 2009
|
|
7VFL
 
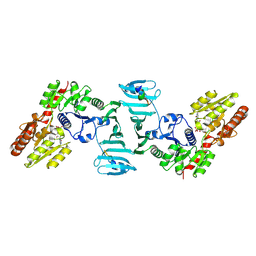 | | Crystal structure of SdgB (UDP, NAG, and O-glycosylated SD peptide-binding form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, group 1 family protein, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2LTJ
 
 | | Conformational analysis of StrH, the surface-attached exo- beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae | | Descriptor: | Beta-N-acetylhexosaminidase | | Authors: | Pluvinage, B, Chitayat, S, Ficko-Blean, E, Abbott, D, Kunjachen, J, Grondin, J, Spencer, H, Smith, S, Boraston, A. | | Deposit date: | 2012-05-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational analysis of StrH, the surface-attached exo-beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae.
J.Mol.Biol., 425, 2013
|
|
4MLL
 
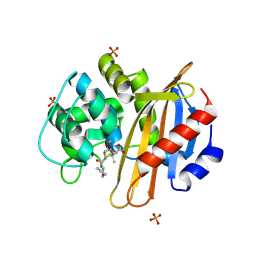 | | The 1.4 A structure of the class D beta-lactamase OXA-1 K70D complexed with oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase OXA-1, ... | | Authors: | June, C.M, Vallier, B.C, Bonomo, R.A, Leonard, D.A, Powers, R.A. | | Deposit date: | 2013-09-06 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural Origins of Oxacillinase Specificity in Class D beta-Lactamases.
Antimicrob.Agents Chemother., 58, 2014
|
|
1RBJ
 
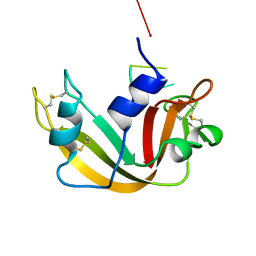 | |
4IMN
 
 | | Crystal structure of wild type human Lipocalin PGDS bound with PEG MME 2000 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Lipocalin-type prostaglandin-D synthase | | Authors: | Lim, S.M, Chen, D, Teo, H, Roos, A, Nyman, T, Tresaugues, L, Pervushin, K, Nordlund, P. | | Deposit date: | 2013-01-03 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and dynamic insights into substrate binding and catalysis of human lipocalin prostaglandin D synthase.
J.Lipid Res., 54, 2013
|
|
110D
 
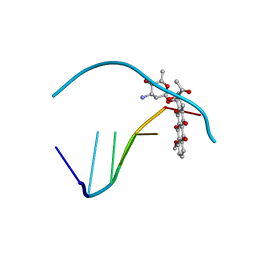 | | ANTHRACYCLINE-DNA INTERACTIONS AT UNFAVOURABLE BASE BASE-PAIR TRIPLET-BINDING SITES: STRUCTURES OF D(CGGCCG)/DAUNOMYCIN AND D(TGGCCA)/ADRIAMYCIN COMPL | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*GP*CP*CP*G)-3') | | Authors: | Leonard, G.A, Hambley, T.W, McAuley-Hecht, K, Brown, T, Hunter, W.N. | | Deposit date: | 1993-01-21 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anthracycline-DNA interactions at unfavourable base-pair triplet-binding sites: structures of d(CGGCCG)/daunomycin and d(TGGCCA)/adriamycin complexes.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
7ZWH
 
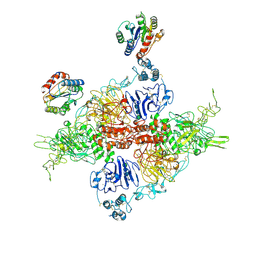 | | VWF Tubules of D1D2 and D'D3A1 domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2022-05-19 | | Release date: | 2022-07-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Assembly of von Willebrand factor tubules with in vivo helical parameters requires A1 domain insertion.
Blood, 140, 2022
|
|
2VVQ
 
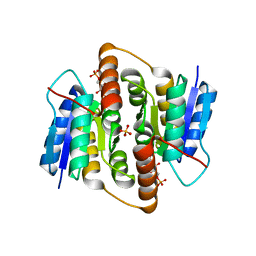 | | Crystal structure of Mycobacterium tuberculosis ribose-5-phosphate isomerase B in complex with the inhibitor 5-deoxy-5-phospho-D- ribonate | | Descriptor: | 5-O-phosphono-D-ribonic acid, RIBOSE-5-PHOSPHATE ISOMERASE B, SULFATE ION | | Authors: | Kowalinski, E, Roos, A.K, Mariano, S, Salmon, L, Mowbray, S.L. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-Ribose-5-Phosphate Isomerase B from Escherichia Coli is Also a Functional D-Allose-6-Phosphate Isomerase, While the Mycobacterium Tuberculosis Enzyme is not.
J.Mol.Biol., 382, 2008
|
|
6AGM
 
 | |
4AZ7
 
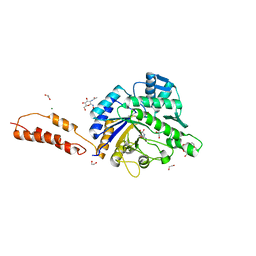 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-23 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
1D42
 
 | |
5YX1
 
 | | Crystal structure of hematopoietic prostaglandin D synthase in complex with U004 | | Descriptor: | GLUTATHIONE, GLYCEROL, Hematopoietic prostaglandin D synthase, ... | | Authors: | Kamo, M, Furubayashi, N, Inaka, K, Aritake, K, Urade, Y, Takaya, D, Tanaka, A. | | Deposit date: | 2017-12-01 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure of hematopoietic prostaglandin D synthase in complex with U004
To Be Published
|
|
201D
 
 | |
4OS0
 
 | | Three-dimensional structure of the C65A-W54F double mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6KNC
 
 | | PolD-PCNA-DNA (form B) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
5AQ0
 
 | | The structure of the Transthyretin-like domain of the first catalytic domain of the HUMAN Carboxypeptidase D | | Descriptor: | CARBOXYPEPTIDASE D, GLYCEROL | | Authors: | Gallego, P, Garcia-Pardo, J, Lorenzo, J, Aviles, F.X, Ventura, S, Reverter, D. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The Structure of the Ttldomain of the Human Carboxypeptidase D
To be Published
|
|
