7R4N
 
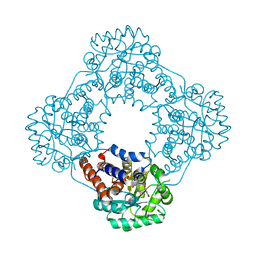 | | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-N-methyl-1H-indazole-3-carboxamide, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Mackinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandeo-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2022-02-08 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide
To Be Published
|
|
3NVS
 
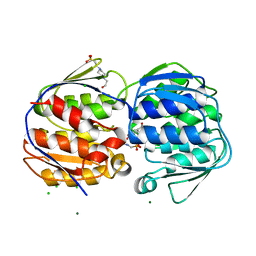 | | 1.02 Angstrom resolution crystal structure of 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae in complex with shikimate-3-phosphate (partially photolyzed) and glyphosate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, G, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.021 Å) | | Cite: | 1.02 Angstrom Resolution Crystal Structure of 3-Phosphoshikimate 1-Carboxyvinyltransferase from Vibrio cholerae in complex with Shikimate-3-Phosphate (Partially Photolyzed) and Glyphosate
TO BE PUBLISHED
|
|
5HP1
 
 | |
8WLA
 
 | | Cryo-EM structure of the beta-1,3-glucan synthase FKS1-Rho1 complex | | Descriptor: | 1,3-beta-glucan synthase component FKS1, GTP-binding protein RHO1 | | Authors: | Li, J.L, Zhu, A.Q, Liu, J.X, Dai, X.L, Yan, C.Y, Deng, D, Wang, X. | | Deposit date: | 2023-09-29 | | Release date: | 2025-02-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the beta-1,3-glucan synthase FKS1-Rho1 complex.
Nat Commun, 16, 2025
|
|
8WL6
 
 | | Structure of 1,3-beta-glucan synthase component FKS1 | | Descriptor: | 1,3-beta-glucan synthase component FKS1 | | Authors: | Li, J.L, Zhu, A.Q, Liu, J.X, Dai, X.L, Wang, X, Yan, C.Y, Deng, D. | | Deposit date: | 2023-09-29 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structure of the beta-1,3-glucan synthase FKS1-Rho1 complex.
Nat Commun, 16, 2025
|
|
5IA9
 
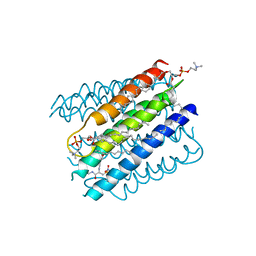 | | The structure of microsomal glutathione transferase 1 in complex with Meisenheimer complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, Microsomal glutathione S-transferase 1, ... | | Authors: | Kuang, Q, Purhonen, P, Jegerschold, C, Morgenstern, R, Hebert, H. | | Deposit date: | 2016-02-21 | | Release date: | 2017-07-12 | | Last modified: | 2025-05-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Dead-end complex, lipid interactions and catalytic mechanism of microsomal glutathione transferase 1, an electron crystallography and mutagenesis investigation.
Sci Rep, 7, 2017
|
|
9EOV
 
 | | Crystal structure of domains I and II from the outer membrane cytochrome MtrC | | Descriptor: | 1,2-ETHANEDIOL, Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC, HEME C, ... | | Authors: | Nash, B.W, Morales Florez, A, Lockwood, C.W.J, Edwards, M.J, Butt, J.N, Clarke, T.A. | | Deposit date: | 2024-03-15 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Extracellular catalysis of environmental substrates by Shewanella oneidensis MR-1 occurs via active sites on the C-terminal domains of MtrC.
Protein Sci., 34, 2025
|
|
6S6W
 
 | | Crystal Structure of human ALDH1A3 in complex with 2,6-diphenylimidazo[1,2-a]pyridine (compound GA11) and NAD+ | | Descriptor: | 2,6-diphenylimidazo[1,2-a]pyridine, Aldehyde dehydrogenase family 1 member A3, GLYCEROL, ... | | Authors: | Gelardi, E.L.M, Quattrini, L, LaMotta, C, Ferraris, D.M, Garavaglia, S. | | Deposit date: | 2019-07-03 | | Release date: | 2020-04-22 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Imidazo[1,2-a]pyridine Derivatives as Aldehyde Dehydrogenase Inhibitors: Novel Chemotypes to Target Glioblastoma Stem Cells.
J.Med.Chem., 63, 2020
|
|
5JMP
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC57 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-29 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
6HMH
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-glucosamine) and alpha-1,2-mannobiose | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Lu, D, Zhu, S, Bernardo-Seisdedos, G, Millet, O, Zhang, Y, Sollogoub, M, Jimenez-Barbero, J, Davies, G.J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | From 1,4-Disaccharide to 1,3-Glycosyl Carbasugar: Synthesis of a Bespoke Inhibitor of Family GH99 Endo-alpha-mannosidase.
Org.Lett., 20, 2018
|
|
5JC1
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC55 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-14 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
4M81
 
 | | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with 1-fluoro-alpha-D-glucopyranoside (donor) and p-nitrophenyl beta-D-glucopyranoside (acceptor) at 1.86A resolution | | Descriptor: | 4-nitrophenyl beta-D-glucopyranoside, EXO-1,3-BETA-GLUCANASE, GLYCEROL, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Larsen, D.S, Cutfield, J.F. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Major Change in Regiospecificity for the Exo-1,3-beta-glucanase from Candida albicans following Its Conversion to a Glycosynthase.
Biochemistry, 53, 2014
|
|
5JBI
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC52 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-13 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
5JNL
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC54 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-30 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
5JO0
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC56 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
6GWR
 
 | | Structure of the kinase domain of human DDR1 in complex with a potent and selective inhibitor of DDR1 and DDR2 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-benzamide, Epithelial discoidin domain-containing receptor 1, ... | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 3-(Imidazo[1,2- a]pyrazin-3-ylethynyl)-4-isopropyl- N-(3-((4-methylpiperazin-1-yl)methyl)-5-(trifluoromethyl)phenyl)benzamide as a Dual Inhibitor of Discoidin Domain Receptors 1 and 2.
J. Med. Chem., 61, 2018
|
|
7XMG
 
 | | Cryo-EM structure of human NaV1.7/beta1/beta2-TCN-1752 | | Descriptor: | (1~{Z})-~{N}-[2-methyl-3-[(~{E})-[6-[4-[[4-(trifluoromethyloxy)phenyl]methoxy]piperidin-1-yl]-1~{H}-1,3,5-triazin-2-ylidene]amino]phenyl]ethanimidic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7WYJ
 
 | | Structure of the complex of lactoperoxidase with nitric oxide catalytic product nitrite at 1.89 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Pandey, N, Singh, A.K, Sinha, M, Singh, R.P, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural evidence of the conversion of nitric oxide (NO) to nitrite ion (NO2-) by lactoperoxidase (LPO): Structure of the complex of LPO with NO2- at 1.89 angstrom resolution
J.Inorg.Biochem., 247, 2023
|
|
5HRO
 
 | |
6IKA
 
 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:entecavir-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6IK9
 
 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
8TDB
 
 | | Structure of PYCR1 complexed with NADH and 1-hydroxyethane-1-sulfonate | | Descriptor: | (1R)-1-hydroxyethane-1-sulfonic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
7X3Z
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 1 complexed with estrone and NAD | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, 17-beta-hydroxysteroid dehydrogenase type 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, T, Lin, S.X, Yin, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New insights into the substrate inhibition of human 17 beta-hydroxysteroid dehydrogenase type 1.
J.Steroid Biochem.Mol.Biol., 228, 2023
|
|
6HMG
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-glucosamine) | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Lu, D, Zhu, S, Bernardo-Seisdedos, G, Millet, O, Zhang, Y, Sollogoub, M, Jimenez-Barbero, J, Davies, G.J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | From 1,4-Disaccharide to 1,3-Glycosyl Carbasugar: Synthesis of a Bespoke Inhibitor of Family GH99 Endo-alpha-mannosidase.
Org.Lett., 20, 2018
|
|
6HAK
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with a double stranded RNA represents the RT transcription initiation complex prior to nucleotide incorporation | | Descriptor: | Gag-Pol polyprotein, MAGNESIUM ION, RNA (5'-R(P*AP*GP*UP*GP*GP*CP*GP*GP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2018-08-07 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of HIV-1 RT/dsRNA initiation complex prior to nucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
