3GRV
 
 | |
5K0R
 
 | | Crystal structure of reduced Shewanella Yellow Enzyme 4 (SYE4) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H:flavin oxidoreductase Sye4, Octadecane | | Authors: | Elegheert, J, Brige, A, Savvides, S.N. | | Deposit date: | 2016-05-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural dissection of Shewanella oneidensis old yellow enzyme 4 bound to a Meisenheimer complex and (nitro)phenolic ligands.
FEBS Lett., 591, 2017
|
|
2OVE
 
 | | Crystal Structure of Recombinant Human Complement Protein C8gamma | | Descriptor: | Complement component 8, gamma polypeptide | | Authors: | Chiswell, B, Lovelace, L.L, Brannen, C, Ortlund, E.A, Lebioda, L, Sodetz, J.M. | | Deposit date: | 2007-02-13 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural features of the ligand binding site on human complement protein C8gamma: A member of the lipocalin family
Biochim.Biophys.Acta, 1774, 2007
|
|
5Y31
 
 | | Crystal structure of human LGI1-ADAM22 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 22, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2017-07-27 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (7.125 Å) | | Cite: | Structural basis of epilepsy-related ligand-receptor complex LGI1-ADAM22.
Nat Commun, 9, 2018
|
|
3GRY
 
 | |
5K1U
 
 | |
2I19
 
 | | T. Brucei farnesyl diphosphate synthase complexed with bisphosphonate | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [2-(PYRIDIN-2-YLAMINO)ETHANE-1,1-DIYL]BIS(PHOSPHONIC ACID) | | Authors: | Cao, R, Mao, J, Gao, Y, Robinson, H, Odeh, S, Goddard, A, Oldfield, E. | | Deposit date: | 2006-08-13 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Solid-state NMR, crystallographic, and computational investigation of bisphosphonates and farnesyl diphosphate synthase-bisphosphonate complexes.
J.Am.Chem.Soc., 128, 2006
|
|
9BWF
 
 | | Crystal structure of cellulose oxidative enzyme without ligand | | Descriptor: | COPPER (II) ION, Cellulose oxidative enzyme | | Authors: | Morais, M.A.B, Santos, C.A, Araujo, E.A, Santos, C.R, Morao, L.G, Motta, M.L, Murakami, M.T. | | Deposit date: | 2024-05-21 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A metagenomic 'dark matter' enzyme catalyses oxidative cellulose conversion.
Nature, 639, 2025
|
|
4EE4
 
 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with tetrasaccharide from Lacto-N-neohexose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
4EEO
 
 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with GLCNAC-BETA1,6-GlcNAc-ALPHA-benzyl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-benzyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
3O2U
 
 | | S. cerevisiae Ubc12 | | Descriptor: | GLYCEROL, NEDD8-conjugating enzyme UBC12 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
2V0E
 
 | |
5E7Q
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis | | Descriptor: | GLYCEROL, SULFATE ION, acyl-CoA synthetase | | Authors: | Osipiuk, J, Cuff, M.E, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-12 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
4WQP
 
 | |
1UMD
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methyl-2-oxopentanoate as an intermediate | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
2OVA
 
 | | X-ray structure of Human Complement Protein C8gamma Y83W Mutant | | Descriptor: | Complement component 8, gamma polypeptide | | Authors: | Chiswell, B, Lovelace, L.L, Brannen, C, Ortlund, E.A, Lebioda, L, Sodetz, J.M. | | Deposit date: | 2007-02-13 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural features of the ligand binding site on human complement protein C8gamma: A member of the lipocalin family
Biochim.Biophys.Acta, 1774, 2007
|
|
2V0F
 
 | |
4EY8
 
 | | Crystal structure of recombinant human acetylcholinesterase in complex with fasciculin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, Fasciculin-2, ... | | Authors: | Cheung, J, Rudolph, M, Burshteyn, F, Cassidy, M, Gary, E, Love, J, Height, J, Franklin, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5958 Å) | | Cite: | Structures of human acetylcholinesterase in complex with pharmacologically important ligands.
J.Med.Chem., 55, 2012
|
|
2I8A
 
 | |
4EE5
 
 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with trisaccharide from Lacto-N-neotetraose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
4EEM
 
 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with GLCNAC-BETA1,6-MAN-ALPHA-methyl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-methyl alpha-D-mannopyranoside, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
1IXP
 
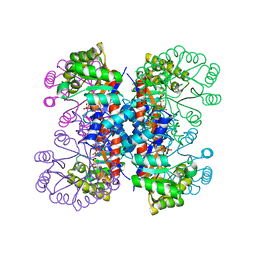 | | Enzyme-phosphate Complex of Pyridoxine 5'-Phosphate synthase | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-Phosphate synthase | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
2II3
 
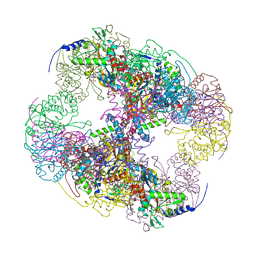 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Oxidized Coenzyme A-bound form | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
2II4
 
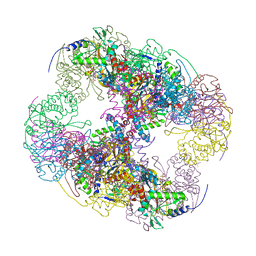 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Coenzyme A-bound form | | Descriptor: | CHLORIDE ION, COENZYME A, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
4EY6
 
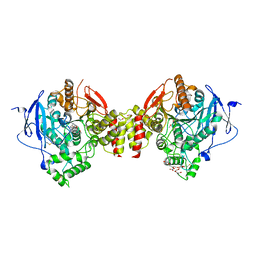 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with (-)-galantamine | | Descriptor: | (-)-GALANTHAMINE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheung, J, Rudolph, M, Burshteyn, F, Cassidy, M, Gary, E, Love, J, Height, J, Franklin, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3983 Å) | | Cite: | Structures of human acetylcholinesterase in complex with pharmacologically important ligands.
J.Med.Chem., 55, 2012
|
|
