2R15
 
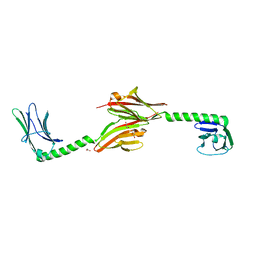 | |
2W70
 
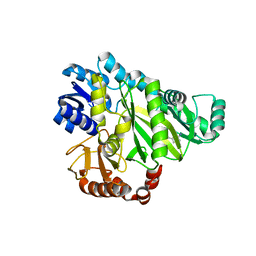 | |
2W6Q
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with the triazine-2,4-diamine fragment | | Descriptor: | 6-(2-phenoxyethoxy)-1,3,5-triazine-2,4-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
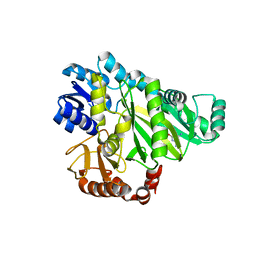
2W71
 
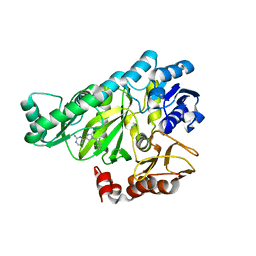 | |
2AUH
 
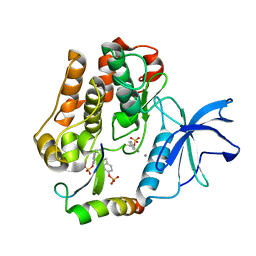 | | Crystal structure of the Grb14 BPS region in complex with the insulin receptor tyrosine kinase | | Descriptor: | CALCIUM ION, Growth factor receptor-bound protein 14, Insulin receptor | | Authors: | Depetris, R.S, Hu, J, Gimpelevich, I, Holt, L.J, Daly, R.J, Hubbard, S.R. | | Deposit date: | 2005-08-27 | | Release date: | 2005-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of the insulin receptor by the adaptor protein grb14.
Mol.Cell, 20, 2005
|
|
2WEX
 
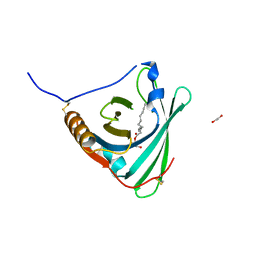 | | Crystal structure of human apoM in complex with glycerol 1- myristic acid | | Descriptor: | (2R)-2,3-dihydroxypropyl tetradecanoate, 1,2-ETHANEDIOL, APOLIPOPROTEIN M | | Authors: | Sevvana, M, Ahnstrom, J, Egerer-Sieber, C, Dahlback, B, Muller, Y.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serendipitous Fatty Acid Binding Reveals the Structural Determinants for Ligand Recognition in Apolipoprotein M.
J.Mol.Biol., 393, 2009
|
|
2B81
 
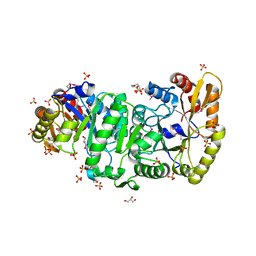 | | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Luciferase-like monooxygenase, ... | | Authors: | Kim, Y, Li, H, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-06 | | Release date: | 2005-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus
To be Published
|
|
6BHT
 
 | | HIV-1 CA hexamer in complex with IP6, orthorhombic crystal form | | Descriptor: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
2BAK
 
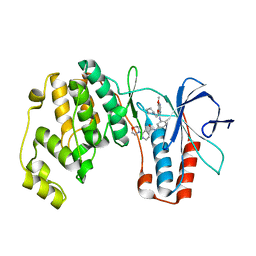 | | p38alpha MAP kinase bound to MPAQ | | Descriptor: | Mitogen-activated protein kinase 14, N-(3-{[7-METHOXY-6-(2-PYRROLIDIN-1-YLETHOXY)QUINAZOLIN-4-YL]AMINO}-4-METHYLPHENYL)-2-MORPHOLIN-4-YLISONICOTINAMIDE | | Authors: | Gerhardt, S, Breed, J, Pauptit, R.A, Read, J, Norman, R.A, Ward, W.H. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase
Biochemistry, 44, 2005
|
|
2BAJ
 
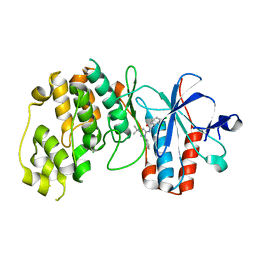 | | p38alpha bound to pyrazolourea | | Descriptor: | 1-(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)-3-(2,3-dichlorophenyl)urea, Mitogen-activated protein kinase 14 | | Authors: | Gerhardt, S, Pauptit, R.A, Read, J, Breed, J, Norman, R.A, Ward, W.H. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase
Biochemistry, 44, 2005
|
|
2BAQ
 
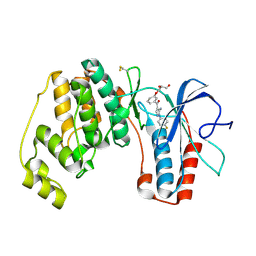 | | p38alpha bound to Ro3201195 | | Descriptor: | Mitogen-activated protein kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE | | Authors: | Gerhardt, S, Pauptit, R.A, Breed, J, Read, J, Tucker, J, Norman, R.A. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase.
Biochemistry, 44, 2005
|
|
2QG4
 
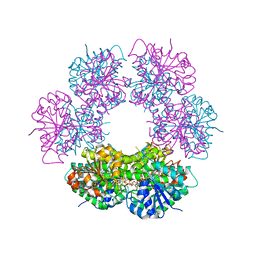 | | Crystal structure of human UDP-glucose dehydrogenase product complex with UDP-glucuronate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kavanagh, K.L, Guo, K, Bunkoczi, G, Savitsky, P, Pilka, E, Bhatia, C, Niesen, F, Smee, C, Berridge, G, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
6BHS
 
 | | HIV-1 CA hexamer in complex with IP6, hexagonal crystal form | | Descriptor: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
2X34
 
 | | Structure of a polyisoprenoid binding domain from Saccharophagus degradans implicated in plant cell wall breakdown | | Descriptor: | CELLULOSE-BINDING PROTEIN, X158, Ubiquinone-8 | | Authors: | Vincent, F, Dal Molin, D, Weiner, R.M, Bourne, Y, Henrissat, B. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-23 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Polyisoprenoid Binding Domain from Saccharophagus Degradans Implicated in Plant Cell Wall Breakdown
FEBS Lett., 584, 2010
|
|
2X31
 
 | | Modelling of the complex between subunits BchI and BchD of magnesium chelatase based on single-particle cryo-EM reconstruction at 7.5 ang | | Descriptor: | MAGNESIUM-CHELATASE 38 KDA SUBUNIT, MAGNESIUM-CHELATASE 60 KDA SUBUNIT | | Authors: | Lunqvist, J, Elmlund, H, Peterson Wulff, R, Berglund, L, Elmlund, D, Emanuelsson, C, Hebert, H, Willows, R.D, Hansson, M, Lindahl, M, Al-Karadaghi, S. | | Deposit date: | 2010-01-19 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | ATP-Induced Conformational Dynamics in the Aaa+ Motor Unit of Magnesium Chelatase.
Structure, 18, 2010
|
|
2PTD
 
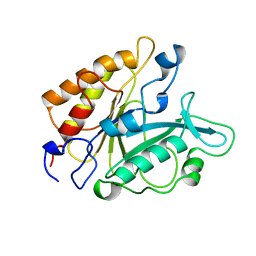 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT D198E | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-16 | | Release date: | 1998-01-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
2QA1
 
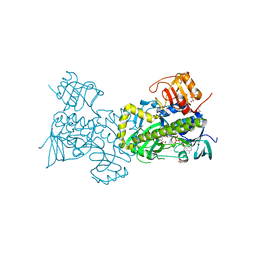 | | Crystal structure of PgaE, an aromatic hydroxylase involved in angucycline biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
2QA2
 
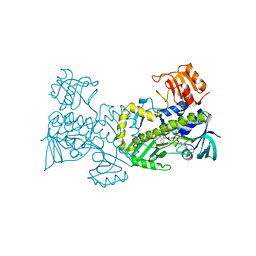 | | Crystal structure of CabE, an aromatic hydroxylase from angucycline biosynthesis, determined to 2.7 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyketide oxygenase CabE | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
2QD9
 
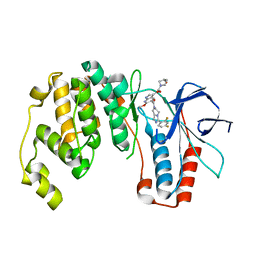 | | P38 Alpha Map Kinase inhibitor based on heterobicyclic scaffolds | | Descriptor: | 1-[5-[[3-[2,4-bis(fluoranyl)phenyl]-6,8-dihydro-5~{H}-imidazo[1,5-a]pyrazin-7-yl]carbonyl]-6-methoxy-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]-2-[(3~{R})-3-oxidanylpyrrolidin-1-yl]ethane-1,2-dione, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and SAR of p38alpha MAP kinase inhibitors based on heterobicyclic scaffolds.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2QJ6
 
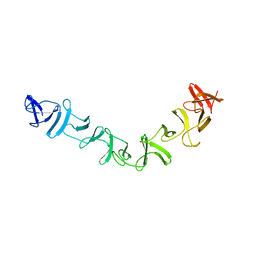 | | Crystal structure analysis of a 14 repeat C-terminal fragment of toxin TcdA in Clostridium difficile | | Descriptor: | Toxin A | | Authors: | Albesa-Jove, D, Bertrand, T, Carpenter, L, Lim, J, Brown, K.A, Fairweather, N. | | Deposit date: | 2007-07-06 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solution and crystal structures of the cell binding domain of toxins TcdA and TcdB from Clostridium difficile
To be Published
|
|
2WH8
 
 | | Interaction of Mycobacterium tuberculosis CYP130 with heterocyclic arylamines | | Descriptor: | 5-AMINO-2-{4-[(4-AMINOPHENYL)SULFANYL]PHENYL}-1H-ISOINDOLE-1,3(2H)-DIONE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 130 | | Authors: | Podust, L.M, Ouellet, H, von Kries, J.P, Ortiz de Montellano, P.R. | | Deposit date: | 2009-05-01 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction of Mycobacterium tuberculosis CYP130 with heterocyclic arylamines.
J. Biol. Chem., 284, 2009
|
|
6B1O
 
 | | The structure of DPP4 in complex with Vildagliptin Analog | | Descriptor: | (2S)-2-amino-1-[(1S,3S,5S)-3-(aminomethyl)-2-azabicyclo[3.1.0]hexan-2-yl]-2-[(1r,3R,5S,7S)-3,5-dihydroxytricyclo[3.3.1.1~3,7~]decan-1-yl]ethan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2017-09-18 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A comparative study of the binding properties, dipeptidyl peptidase-4 (DPP-4) inhibitory activity and glucose-lowering efficacy of the DPP-4 inhibitors alogliptin, linagliptin, saxagliptin, sitagliptin and vildagliptin in mice.
Endocrinol Diabetes Metab, 1, 2018
|
|
2AYL
 
 | | 2.0 Angstrom Crystal Structure of Manganese Protoporphyrin IX-reconstituted Ovine Prostaglandin H2 Synthase-1 Complexed With Flurbiprofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, GLYCEROL, ... | | Authors: | Gupta, K, Selinsky, B.S, Loll, P.J. | | Deposit date: | 2005-09-07 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 angstroms structure of prostaglandin H2 synthase-1 reconstituted with a manganese porphyrin cofactor.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2Q3E
 
 | | Structure of human UDP-glucose dehydrogenase complexed with NADH and UDP-glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, UDP-glucose 6-dehydrogenase, ... | | Authors: | Kavanagh, K.L, Guo, K, Bunkoczi, G, Savitsky, P, Pilka, E, Bhatia, C, Smee, C, Berridge, G, von Delft, F, Wiegelt, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
2WTB
 
 | | Arabidopsis thaliana multifuctional protein, MFP2 | | Descriptor: | FATTY ACID MULTIFUNCTIONAL PROTEIN (ATMFP2) | | Authors: | Arent, S, Pye, V.E, Christensen, C.E, Norgaard, A, Henriksen, A. | | Deposit date: | 2009-09-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Multi-Functional Protein in Peroxisomal Beta-Oxidation. Structure and Substrate Specificity of the Arabidopsis Thaliana Protein, Mfp2
J.Biol.Chem., 285, 2010
|
|
