8PCE
 
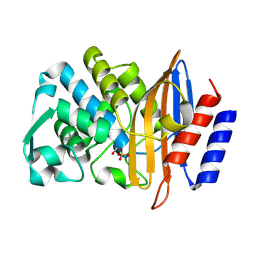 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 250 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
5HLL
 
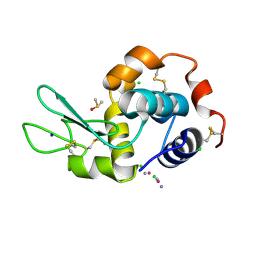 | |
6JQS
 
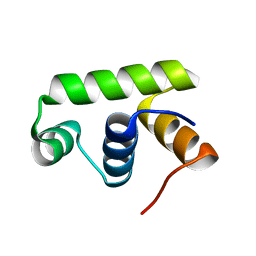 | | Structure of Transcription factor, GerE | | Descriptor: | DNA-binding response regulator | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a transcription factor, GerE (PaGerE), from spore-forming bacterium Paenisporosarcina sp. TG-14.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
3WUU
 
 | |
4BGJ
 
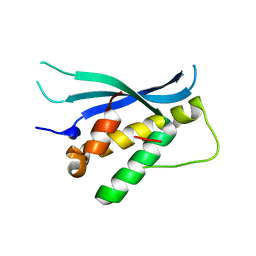 | | Crystal structure of the phox-homology domain of human sorting nexin 14 | | Descriptor: | SORTING NEXIN-14 | | Authors: | Vollmar, M, Kiyani, W, Shrestha, L, Goubin, S, Krojer, T, Pike, A.C.W, Carpenter, E, Quigley, A, McKenzie, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Yue, W.W. | | Deposit date: | 2013-03-27 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Phox-Homology Domain of Human Sorting Nexin 14
To be Published
|
|
3ONT
 
 | |
8EKT
 
 | | CYP51 from Acanthamoeba castellanii in complex with the tetrazole-based IND inhibitor VT-1161(VT1) | | Descriptor: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, PROTOPORPHYRIN IX CONTAINING FE, sterol 14a-demethylase | | Authors: | Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Identification of Potent and Selective Inhibitors of Acanthamoeba : Structural Insights into Sterol 14 alpha-Demethylase as a Key Drug Target.
J.Med.Chem., 67, 2024
|
|
6JJR
 
 | | Crystal structure of human chitotriosidase-1 (hChit1) catalytic domain in complex with compound 2-8-14 | | Descriptor: | 6-azanyl-2-oxidanylidene-N-[(1S)-1-phenylethyl]-7-(phenylmethyl)-1$l^{4},9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-1(14),3(8),4,6,10,12-hexaene-5-carboxamide, Chitotriosidase-1 | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-02-26 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6SO0
 
 | | NMR solution structure of the family 14 carbohydrate binding module (CBM14) from human chitotriosidase | | Descriptor: | Chitotriosidase-1 | | Authors: | Madland, E, Crasson, O, Vandevenne, M, Sorlie, M, Aachmann, F.L. | | Deposit date: | 2019-08-28 | | Release date: | 2020-01-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR and Fluorescence Spectroscopies Reveal the Preorganized Binding Site in Family 14 Carbohydrate-Binding Module from Human Chitotriosidase.
Acs Omega, 4, 2019
|
|
6MCW
 
 | | Crystal structure of the P450 domain of the CYP51-ferredoxin fusion protein from Methylococcus capsulatus, complex with the detergent Anapoe-X-114 | | Descriptor: | 23-[4-(2,4,4-trimethylpentan-2-yl)phenoxy]-3,6,9,12,15,18,21-heptaoxatricosan-1-ol, Cytochrome P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T, Wawrzak, Z, Lamb, D.C, Lepesheva, G.I. | | Deposit date: | 2018-09-02 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Concerning P450 evolution: Structural Analyses Support Bacterial Origin of Sterol 14 alpha-Demethylases.
Mol.Biol.Evol., 2020
|
|
6MI0
 
 | | Crystal structure of the P450 domain of the CYP51-ferredoxin fusion protein from Methylococcus capsulatus, ligand-free state | | Descriptor: | Cytochrome P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T, Wawrzak, Z, Lamb, D.C, Lepesheva, G.I. | | Deposit date: | 2018-09-18 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Concerning P450 evolution: Structural Analyses Support Bacterial Origin of Sterol 14 alpha-Demethylases.
Mol.Biol.Evol., 2020
|
|
4UYL
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51B) from a pathogenic filamentous fungus Aspergillus fumigatus in complex with VNI | | Descriptor: | 14-ALPHA STEROL DEMETHYLASE, N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2014-09-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure-Functional Characterization of Cytochrome P450 Sterol Alpha-Demethylase (Cyp51B) from Aspergillus Fumigatus and Molecular Basis for the Development of Antifungal Drugs
J.Biol.Chem., 290, 2015
|
|
4V52
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with neomycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8HYJ
 
 | |
4V5A
 
 | | Structure of the Ribosome Recycling Factor bound to the Thermus thermophilus 70S ribosome with mRNA, ASL-Phe and tRNA-fMet | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Petry, S, Dunham, C.M, Selmer, M, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2007-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the ribosome recycling factor bound to the ribosome.
Nat. Struct. Mol. Biol., 14, 2007
|
|
6TCH
 
 | |
6DDD
 
 | | Structure of the 50S ribosomal subunit from Methicillin Resistant Staphylococcus aureus in complex with the oxazolidinone antibiotic LZD-5 | | Descriptor: | 2,2-dichloro-N-({(5S)-3-[3-fluoro-4-(morpholin-4-yl)phenyl]-2-oxo-1,3-oxazolidin-5-yl}methyl)acetamide, 23S rRNA, 50S ribosomal protein L13, ... | | Authors: | Belousoff, M.J, Venugopal, H, Bamert, R.S, Lithgow, T. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | cryoEM-Guided Development of Antibiotics for Drug-Resistant Bacteria.
ChemMedChem, 14, 2019
|
|
6DDG
 
 | | Structure of the 50S ribosomal subunit from Methicillin Resistant Staphylococcus aureus in complex with the oxazolidinone antibiotic LZD-6 | | Descriptor: | 2-chloro-N-({(5S)-3-[3-fluoro-4-(morpholin-4-yl)phenyl]-2-oxo-1,3-oxazolidin-5-yl}methyl)acetamide, 23S rRNA, 50S ribosomal protein L13, ... | | Authors: | Belousoff, M.J, Venugopal, H, Bamert, R.S, Lithgow, T. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | cryoEM-Guided Development of Antibiotics for Drug-Resistant Bacteria.
ChemMedChem, 14, 2019
|
|
6T7H
 
 | | Crystal structure of Thrombin in complex with macrocycle N14-PR4-A | | Descriptor: | (14S,17R)-14-(3-carbamimidamidopropyl)-3-(furan-2-ylmethyl)-5,12,15-tris(oxidanylidene)-19-thia-3,6,13,16-tetrazatricyclo[19.4.0.0^{6,10}]pentacosa-1(25),7,9,21,23-pentaene-17-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Angelini, A, Kumar, M.G, Heinis, C, Cendron, L. | | Deposit date: | 2019-10-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Macrocycle synthesis strategy based on step-wise "adding and reacting" three components enables screening of large combinatorial libraries.
Chem Sci, 11, 2020
|
|
1Y1A
 
 | | CRYSTAL STRUCTURE OF CALCIUM AND INTEGRIN BINDING PROTEIN | | Descriptor: | CALCIUM ION, Calcium and integrin binding 1 (calmyrin), GLUTATHIONE | | Authors: | Blamey, C.J, Ceccarelli, C, Naik, U.P, Bahnson, B.J. | | Deposit date: | 2004-11-17 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of calcium- and integrin-binding protein 1: Insights into redox regulated functions
Protein Sci., 14, 2005
|
|
4V64
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with hygromycin B. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Shoji, S, Fredrick, K, Cate, J.H.D. | | Deposit date: | 2008-06-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for hygromycin B inhibition of protein biosynthesis
Rna, 14, 2008
|
|
7O1D
 
 | |
8C83
 
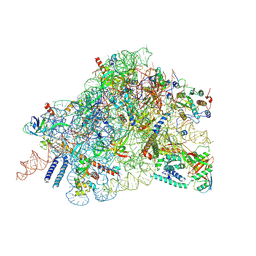 | | Cryo-EM structure of in vitro reconstituted Otu2-bound Ub-40S complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
7O2L
 
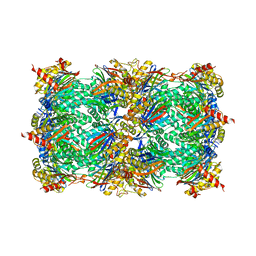 | | Yeast 20S proteasome in complex with the covalently bound inhibitor b-lactone (2R,3S)-3-isopropyl-4-oxo-2-oxetane-carboxylate (IOC) | | Descriptor: | (2 {R},3 {S})-3-methanoyl-4-methyl-2-hydroxy-pentanoic acid, 20S proteasome, BJ4_G0020160.mRNA.1.CDS.1, ... | | Authors: | Shi, Y.M, Hirschmann, M, Shi, Y.N, Shabbir, A, Abebew, D, Tobias, N.J, Gruen, P, Crames, J.J, Poeschel, L, Kuttenlochner, W, Richter, C, Herrmann, J, Mueller, R, Thanwisai, A, Pidot, S.J, Stinear, T.P, Groll, M, Kim, Y, Bode, H. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Nat.Chem., 14, 2022
|
|
5WBS
 
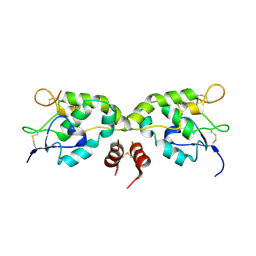 | | Crystal structure of Frizzled-7 CRD with an inhibitor peptide Fz7-21 | | Descriptor: | Frizzled-7,inhibitor peptide Fz7-21 | | Authors: | Nile, A.H, Mukund, S, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
