1JLU
 
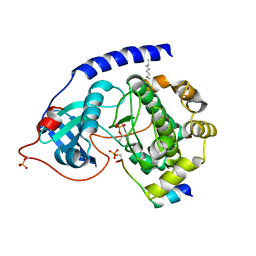 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Phosphorylated Substrate Peptide and Detergent | | Descriptor: | AMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.-H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
5VNI
 
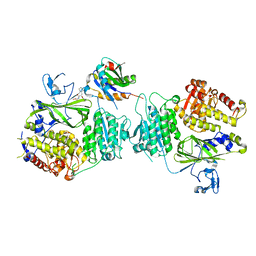 | |
3L9N
 
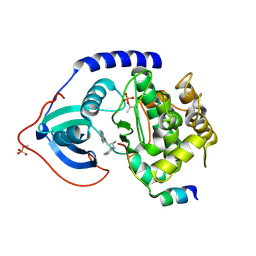 | | crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 27 | | Descriptor: | (2S)-N~1~-[5-(1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L9L
 
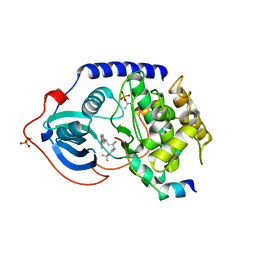 | | Crystal structure of pka with compound 36 | | Descriptor: | 5-[2-({(2S)-2-amino-3-[4-(trifluoromethyl)phenyl]propyl}amino)-1,3-thiazol-5-yl]-1,3-dihydro-2H-indol-2-one, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XWB
 
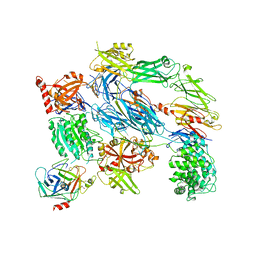 | | Crystal Structure of Complement C3b in complex with Factors B and D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3B ALPHA' CHAIN, ... | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
2Y48
 
 | | Crystal structure of LSD1-CoREST in complex with a N-terminal SNAIL peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | Authors: | Baron, R, Binda, C, Tortorici, M, McCammon, J.A, Mattevi, A. | | Deposit date: | 2011-01-05 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Mimicry and Ligand Recognition in Binding and Catalysis by the Histone Demethylase Lsd1-Corest Complex.
Structure, 19, 2011
|
|
2Y5B
 
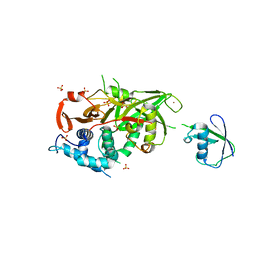 | | Structure of USP21 in complex with linear diubiquitin-aldehyde | | Descriptor: | SULFATE ION, UBIQUITIN, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 21, ... | | Authors: | Ye, Y, Akutsu, M, Reyes-Turcu, F, Enchev, R.I, Wilkinson, K.D, Komander, D. | | Deposit date: | 2011-01-12 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Polyubiquitin Binding and Cross-Reactivity in the Usp Domain Deubiquitinase Usp21.
Embo Rep., 12, 2011
|
|
2UV2
 
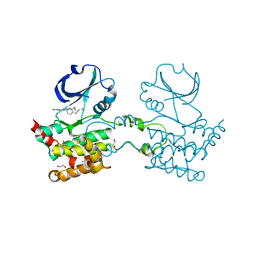 | | Crystal Structure Of Human Ste20-Like Kinase Bound To 4-(4-(5- Cyclopropyl-1H-pyrazol-3-ylamino)-quinazolin-2-ylamino)-phenyl)- acetonitrile | | Descriptor: | 1,2-ETHANEDIOL, STE20-LIKE SERINE-THREONINE KINASE, THIOCYANATE ION, ... | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Keates, T, Salah, E, Savitsky, P, Papagrigoriou, E, Bunkoczi, G, Debreczeni, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-03-08 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
1IRI
 
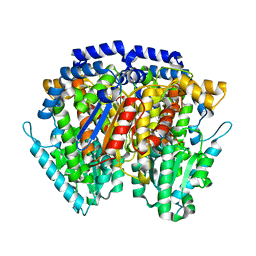 | | Crystal structure of human autocrine motility factor complexed with an inhibitor | | Descriptor: | ERYTHOSE-4-PHOSPHATE, autocrine motility factor | | Authors: | Tanaka, N, Haga, A, Uemura, H, Akiyama, H, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2001-10-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies.
J.Mol.Biol., 318, 2002
|
|
3KXX
 
 | | Structure of the mutant Fibroblast Growth Factor receptor 1 | | Descriptor: | Basic fibroblast growth factor receptor 1 | | Authors: | Bae, J.H, Boggon, T.J, Tome, F, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2009-12-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KLT
 
 | | Crystal structure of a vimentin fragment | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Nicolet, S, Strelkov, S.V. | | Deposit date: | 2009-11-09 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atomic structure of vimentin coil 2.
J.Struct.Biol., 170, 2010
|
|
5VNO
 
 | | Crystal structure of Sec23a/Sec24a/Sec22 | | Descriptor: | Protein transport protein Sec23A, Protein transport protein Sec24A, Vesicle-trafficking protein SEC22b, ... | | Authors: | Ma, W, Goldberg, J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | ER retention is imposed by COPII protein sorting and attenuated by 4-phenylbutyrate.
Elife, 6, 2017
|
|
3LV3
 
 | | Crystal structure of HLA-B*2705 complexed with a peptide derived from the human voltage-dependent calcium channel alpha1 subunit (residues 513-521) | | Descriptor: | 9-meric peptide from Voltage-dependent L-type calcium channel subunit alpha-1D, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Loll, B, Rueckert, C, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2010-02-19 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Loss of recognition by cross-reactive T cells and its relation to a C-terminus-induced conformational reorientation of an HLA-B*2705-bound peptide.
Protein Sci., 20, 2011
|
|
5VNF
 
 | |
5VNL
 
 | |
2YUI
 
 | | Solution structure of the N-terminal domain in human cytokine-induced apoptosis inhibitor anamorsin | | Descriptor: | Anamorsin | | Authors: | Saito, K, Tomizawa, T, Tochio, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain in human cytokine-induced apoptosis inhibitor anamorsin
To be Published
|
|
5VNJ
 
 | |
2YVQ
 
 | | Crystal structure of MGS domain of carbamoyl-phosphate synthetase from homo sapiens | | Descriptor: | Carbamoyl-phosphate synthase | | Authors: | Xie, Y, Ihsanawati, Kishishita, S, Murayama, K, Takemoto, C, Shirozu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of MGS domain of carbamoyl-phosphate synthetase from homo sapiens
To be Published
|
|
2YS4
 
 | | Solution structure of the N-terminal PapD-like domain of HYDIN protein from human | | Descriptor: | Hydrocephalus-inducing protein homolog | | Authors: | Li, H, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal PapD-like domain of HYDIN protein from human
To be Published
|
|
2YRU
 
 | | Solution structure of mouse Steroid receptor RNA activator 1 (SRA1) protein | | Descriptor: | Steroid receptor RNA activator 1 | | Authors: | Nameki, N, Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse Steroid receptor RNA activator 1 (SRA1) protein
To be Published
|
|
1M6D
 
 | | Crystal structure of human cathepsin F | | Descriptor: | 4-MORPHOLIN-4-YL-PIPERIDINE-1-CARBOXYLIC ACID [1-(3-BENZENESULFONYL-1-PROPYL-ALLYLCARBAMOYL)-2-PHENYLETHYL]-AMIDE, Cathepsin F | | Authors: | Somoza, J.R, Palmer, J.T, Ho, J.D. | | Deposit date: | 2002-07-15 | | Release date: | 2003-07-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of human cathepsin F and its implications for the development of novel immunomodulators
J.Mol.Biol., 322, 2002
|
|
1M1L
 
 | | Human Suppressor of Fused (N-terminal domain) | | Descriptor: | Suppressor of Fused | | Authors: | Merchant, M, Vajdos, F.F, Ultsch, M, Maun, H.R, Wendt, U, Cannon, J, Lazarus, R.A, de Vos, A.M, de Sauvage, F.J. | | Deposit date: | 2002-06-19 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Suppressor of fused regulates Gli activity through a dual binding mechanism
Mol.Cell.Biol., 24, 2004
|
|
1MEJ
 
 | | Human Glycinamide Ribonucleotide Transformylase domain at pH 8.5 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
1MEO
 
 | | human glycinamide ribonucleotide Transformylase at pH 4.2 | | Descriptor: | PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase, SULFATE ION | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
2D0W
 
 | | Crystal structure of cytochrome cL from Hyphomicrobium denitrificans | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, cytochrome cL | | Authors: | Nojiri, M, Hira, D, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2005-08-10 | | Release date: | 2006-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of cytochrome c(L) and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins
Biochemistry, 45, 2006
|
|
