8IXS
 
 | | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1/Nrf2 PPI Inhibitor for Treatment of Chronic Kidney Disease | | Descriptor: | (2R,3S)-3-[[(2S)-2-fluoranyl-2-(5,6,7,8-tetrahydronaphthalen-2-yl)ethanoyl]amino]-2-methyl-3-(4-methylphenyl)propanoic acid, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Nomura, A, Yamaguchi, K, Adachi, T. | | Deposit date: | 2023-04-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1-Nrf2 PPI Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
8IVR
 
 | | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1/Nrf2 PPI Inhibitor for Treatment of Chronic Kidney Disease | | Descriptor: | (2R,3S)-2-methyl-3-(4-methylphenyl)-3-[2-(5,6,7,8-tetrahydronaphthalen-2-yl)ethanoylamino]propanoic acid, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Nomura, A, Yamaguchi, K, Adachi, T. | | Deposit date: | 2023-03-28 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1-Nrf2 PPI Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
8JRD
 
 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waki, T, Imaizumi, R, Nakata, S, Yanai, T, Takeshita, K, Sakai, N, Kataoka, K, Yamamoto, M, Nakayama, T, Yamashita, S. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into catalytic promiscuity of chalcone synthase from Glycine max (L.) Merr.: Coenzyme A-induced alteration of product specificity.
Biochem.Biophys.Res.Commun., 718, 2024
|
|
8JYG
 
 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-2-[2-(3-phenoxyphenyl)ethyl]-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based lead optimization to improve potency and selectivity of a novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid series of heparanase-1 inhibitor.
Bioorg.Med.Chem., 93, 2023
|
|
8K5W
 
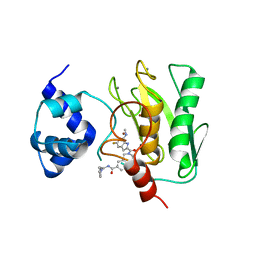 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 2-[[5-fluoranyl-7-(methylamino)-1H-indol-2-yl]carbonyl]-N-(2-pyrrol-1-ylethyl)-3,4-dihydro-1H-isoquinoline-7-carboxamide, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5V
 
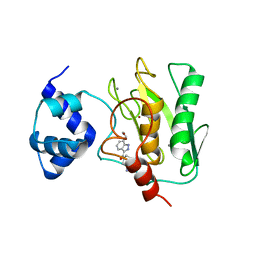 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 6,7-dihydro-4H-[1,3]oxazolo[4,5-c]pyridin-5-yl-(7-ethyl-2H-indazol-3-yl)methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5Y
 
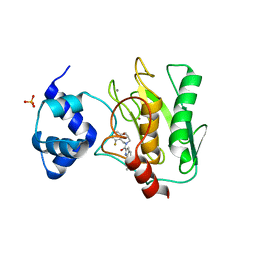 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | (3-azanyl-4-fluoranyl-5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)-[6-(2-oxidanylpropan-2-yl)-1H-indol-2-yl]methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5X
 
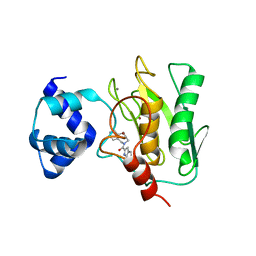 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | (6-cyclopropyl-1~{H}-indol-2-yl)-(5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)methanone, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8IFE
 
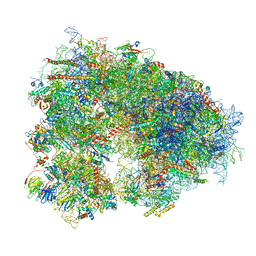 | | Arbekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8IFD
 
 | | Dibekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8IFB
 
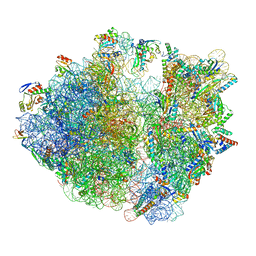 | | Dibekacin-bound E.coli 70S ribosome in the PURE system | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8P2U
 
 | |
2PHB
 
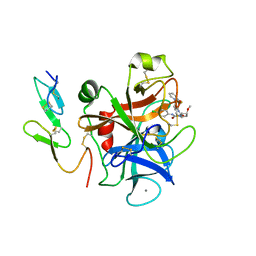 | | An Orally Efficacious Factor Xa Inhibitor | | Descriptor: | (2R,4R)-N~1~-(4-CHLOROPHENYL)-N~2~-[2-FLUORO-4-(2-OXOPYRIDIN-1(2H)-YL)PHENYL]-4-METHOXYPYRROLIDINE-1,2-DICARBOXAMIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Zhang, E, Kohrt, J.T, Bigge, C.F, Finzel, B.C. | | Deposit date: | 2007-04-10 | | Release date: | 2008-03-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The discovery of (2R,4R)-N-(4-chlorophenyl)-N- (2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl)-4-methoxypyrrolidine-1,2-dicarboxamide (PD 0348292), an orally efficacious factor Xa inhibitor
Chem.Biol.Drug Des., 70, 2007
|
|
2Q1J
 
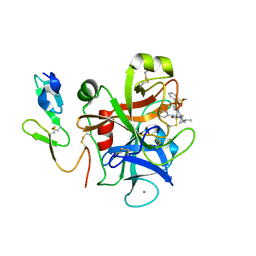 | | The discovery of glycine and related amino acid-based factor xa inhibitors | | Descriptor: | 1-(butyl{[(4-chlorophenyl)amino]carbonyl}amino)-N-[3-fluoro-2'-(methylsulfonyl)biphenyl-4-yl]cyclopropanecarboxamide, Activated factor Xa heavy chain (EC 3.4.21.6), CALCIUM ION, ... | | Authors: | Kohrt, J.T, Filipski, K.J, Cody, W.L, Bigge, C.F, Zhang, E, Finzel, B.C. | | Deposit date: | 2007-05-24 | | Release date: | 2007-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Discovery of Glycine and Related Amino Acid-Based Factor Xa Inhibitors
BIOORG.MED.CHEM., 14, 2006
|
|
8G2O
 
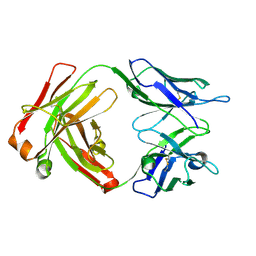 | | Fab structure - Anti-ApoE-7C11 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, anti-ApoE-7C11 heavy chain, anti-ApoE-7C11 light chain | | Authors: | Marino, C, Arboleda-Velasquez, J.F. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | APOE Christchurch-mimetic therapeutic antibody reduces APOE-mediated toxicity and tau phosphorylation.
Alzheimers Dement, 20, 2024
|
|
1IST
 
 | |
5GO1
 
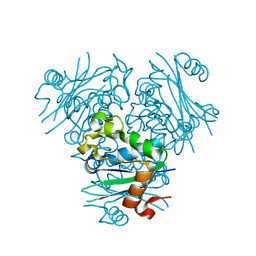 | | Structural, Functional characterization and discovery of novel inhibitors of Leishmania amazonensis Nucleoside Diphosphatase Kinase (NDK) | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Mishra, A.K, Agnihotri, P, Singh, S.P, Pratap, J.V. | | Deposit date: | 2016-07-26 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of novel inhibitors for Leishmania nucleoside diphosphatase kinase (NDK) based on its structural and functional characterization.
J. Comput. Aided Mol. Des., 31, 2017
|
|
8HUB
 
 | | AMP deaminase 2 in complex with an inhibitor | | Descriptor: | 3,3-dimethyl-4-(phenylmethyl)-2~{H}-quinoxaline-1-carboxamide, AMP deaminase 2, ZINC ION | | Authors: | Adachi, T, Doi, S. | | Deposit date: | 2022-12-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The discovery of 3,3-dimethyl-1,2,3,4-tetrahydroquinoxaline-1-carboxamides as AMPD2 inhibitors with a novel mechanism of action.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
8HU6
 
 | | AMP deaminase 2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, AMP deaminase 2, SULFATE ION, ... | | Authors: | Adachi, T, Doi, S. | | Deposit date: | 2022-12-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The discovery of 3,3-dimethyl-1,2,3,4-tetrahydroquinoxaline-1-carboxamides as AMPD2 inhibitors with a novel mechanism of action.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
5GXH
 
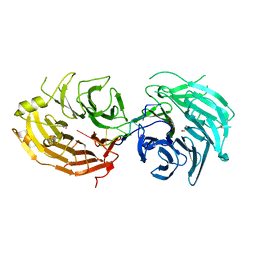 | | The structure of the Gemin5 WD40 domain with AAUUUUUG | | Descriptor: | GLYCEROL, Gem-associated protein 5, RNA (5'-R(*A*AP*UP*UP*UP*UP*UP*G)-3'), ... | | Authors: | Xu, C, He, H, Li, Y, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-17 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly
Genes Dev., 30, 2016
|
|
8H3M
 
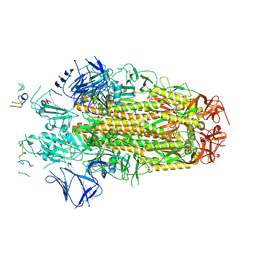 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8H3N
 
 | | Conformation 2 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy-chain, MO1 light chain, ... | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
5GZK
 
 | | Endo-beta-1,2-glucanase from Chitinophaga pinensis - sophorotriose and glucose complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Abe, K, Nakajima, M, Arakawa, T, Fushinobu, S, Taguchi, H. | | Deposit date: | 2016-09-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family
J. Biol. Chem., 292, 2017
|
|
5GZH
 
 | | Endo-beta-1,2-glucanase from Chitinophaga pinensis - ligand free form | | Descriptor: | Endo-beta-1,2-glucanase, IODIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Abe, K, Nakajima, M, Arakawa, T, Fushinobu, S, Taguchi, H. | | Deposit date: | 2016-09-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family
J. Biol. Chem., 292, 2017
|
|
8HV7
 
 | | Crystal structure of EGFR_TMX in complex with covalently bound fragment 9 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DIMETHYL SULFOXIDE, Epidermal growth factor receptor, ... | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
