8AHH
 
 | |
8AHG
 
 | |
8AHE
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHF
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHI
 
 | |
6VDK
 
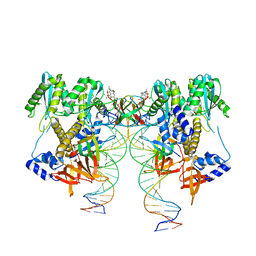 | | CryoEM structure of HIV-1 conserved Intasome Core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Li, M, Chen, X, Craigie, R. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies.
J.Mol.Biol., 432, 2020
|
|
7NBN
 
 | | Allostery through DNA drives phenotype switching | | Descriptor: | AddAB promoter | | Authors: | Rosenblum, G, Elad, N, Rozenberg, H, Wiggers, F, Jungwirth, J, Hofmann, H. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Allostery through DNA drives phenotype switching.
Nat Commun, 12, 2021
|
|
3HOT
 
 | |
2MLK
 
 | | Three-dimensional structure of the C-terminal DNA-binding domain of RstA protein from Klebsiella pneumoniae | | Descriptor: | RstA | | Authors: | Fang, P, Chen, S, Cheng, Y, Chang, C, Yu, T, Huang, T. | | Deposit date: | 2014-03-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element.
Nucleic Acids Res., 42, 2014
|
|
2H9S
 
 | |
1NGN
 
 | | Mismatch repair in methylated DNA. Structure of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4 | | Descriptor: | methyl-CpG binding protein MBD4 | | Authors: | Wu, P, Qiu, C, Sohail, A, Zhang, X, Bhagwat, A.S, Cheng, X. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4
J.Biol.Chem., 278, 2003
|
|
4Z4B
 
 | |
1NZB
 
 | | Crystal structure of wild type Cre recombinase-loxP synapse | | Descriptor: | Cre recombinase, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Meyer, J.E.W, Buchholz, F, Stewart, A.F, Suck, D. | | Deposit date: | 2003-02-17 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation
Nucleic Acids Res., 31, 2003
|
|
3HOS
 
 | |
8WOH
 
 | |
5GN3
 
 | |
2QKK
 
 | | Human RNase H catalytic domain mutant D210N in complex with 14-mer RNA/DNA hybrid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*G)-3', ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Ghirlando, R, Cerritelli, S.M, Crouch, R.J, Yang, W. | | Deposit date: | 2007-07-11 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Mol.Cell, 28, 2007
|
|
1CA5
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
1OUQ
 
 | | Crystal structure of wild-type Cre recombinase-loxP synapse | | Descriptor: | Cre recombinase, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Meyer, J.E.W, Buchholz, F, Stewart, A.F, Suck, D. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation
Nucleic Acids Res., 31, 2003
|
|
1CA6
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
4LTH
 
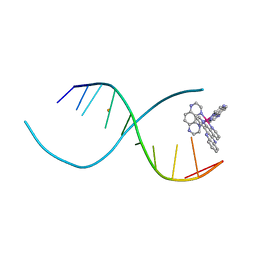 | |
1R0A
 
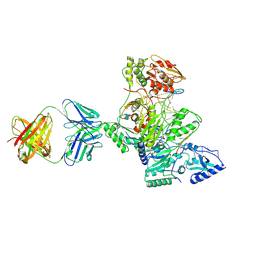 | | Crystal structure of HIV-1 reverse transcriptase covalently tethered to DNA template-primer solved to 2.8 angstroms | | Descriptor: | 5'-D(*A*TP*GP*CP*AP*TP*CP*GP*GP*CP*GP*CP*TP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*GP*GP*T)-3', 5'-D(*C*CP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*AP*GP*CP*GP*CP*CP*GP*(2DA))-3', GLYCEROL, ... | | Authors: | Tuske, S, Ding, J, Arnold, E. | | Deposit date: | 2003-09-19 | | Release date: | 2004-08-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nonnucleoside inhibitor binding affects the interactions of the fingers subdomain of human immunodeficiency virus type 1 reverse transcriptase with DNA.
J.Virol., 78, 2004
|
|
8UJT
 
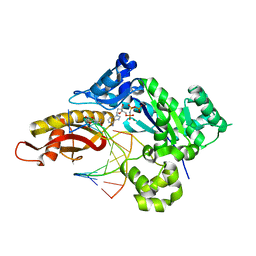 | | Crystal structure of human polymerase eta with incoming dAMPnPP nucleotide opposite urea lesion | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(XB9)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Tomar, R, Egli, M, Stone, M.P. | | Deposit date: | 2023-10-11 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Replication Bypass of the N -(2-Deoxy-d-erythro-pentofuranosyl)-urea DNA Lesion by Human DNA Polymerase eta.
Biochemistry, 63, 2024
|
|
1QUM
 
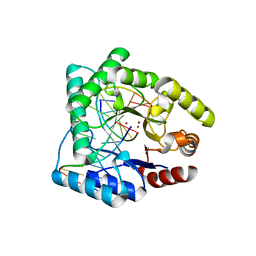 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI ENDONUCLEASE IV IN COMPLEX WITH DAMAGED DNA | | Descriptor: | 5'-D(*(3DR)P*CP*GP*AP*CP*GP*A)-3', 5'-D(*CP*GP*TP*CP*C)-3', 5'-D(*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*G)-3', ... | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-07-01 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
7YVW
 
 | | NMR determination of the 2:1 binding motif structure involving cytosine flipping out for the recognition of the CGG/CGG triad DNA | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3') | | Authors: | Furuita, K, Yamada, T, Sakurabayashi, S, Nomura, M, Kojima, C, Nakatani, K. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.
Nucleic Acids Res., 50, 2022
|
|
