5KCR
 
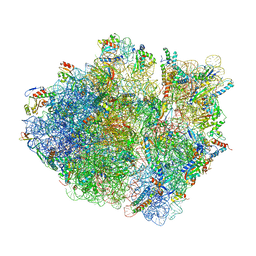 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Avilamycin C, mRNA and P-site tRNA at 3.6A resolution | | Descriptor: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7CHD
 
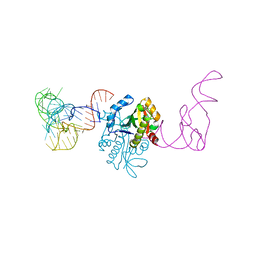 | | AtaT complexed with acetyl-methionyl-tRNAfMet | | Descriptor: | N-acetyltransferase domain-containing protein, RNA (77-MER) | | Authors: | Yashiro, Y, Tomita, K. | | Deposit date: | 2020-07-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.804 Å) | | Cite: | Mechanism of aminoacyl-tRNA acetylation by an aminoacyl-tRNA acetyltransferase AtaT from enterohemorrhagic E. coli.
Nat Commun, 11, 2020
|
|
4QGS
 
 | |
8G3C
 
 | |
4XLR
 
 | |
4XO4
 
 | |
8G3E
 
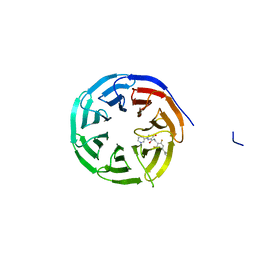 | | Crystal structure of human WDR5 in complex with (1M)-N-[(3,5-difluoro[1,1'-biphenyl]-4-yl)methyl]-6-methyl-4-oxo-1-(pyridin-3-yl)-1,4-dihydropyridazine-3-carboxamide (compound 2, WDR5-MYC inhibitor) | | Descriptor: | (1M)-N-[(3,5-difluoro[1,1'-biphenyl]-4-yl)methyl]-6-methyl-4-oxo-1-(pyridin-3-yl)-1,4-dihydropyridazine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Zhao, M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery and Structure-Based Design of Inhibitors of the WD Repeat-Containing Protein 5 (WDR5)-MYC Interaction.
J.Med.Chem., 66, 2023
|
|
4XP4
 
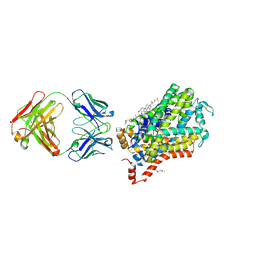 | | X-ray structure of Drosophila dopamine transporter in complex with cocaine | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ANTIBODY FRAGMENT HEAVY CHAIN-PROTEIN, 9D5-HEAVY CHAIN, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
1QMT
 
 | |
4XN8
 
 | |
4XNU
 
 | | X-ray structure of Drosophila dopamine transporter in complex with nisoxetine | | Descriptor: | CHLORIDE ION, CHOLESTEROL, Integral membrane protein-dopamine transporter, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | X-ray structures of Drosophila dopamine transporter in complex with nisoxetine and reboxetine.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XPF
 
 | | X-ray structure of Drosophila dopamine transporter with subsiteB mutations (D121G/S426M) bound to RTI-55 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ANTIBODY FRAGMENT HEAVY CHAIN-PROTEIN, 9D5-HEAVY CHAIN, ... | | Authors: | Penmatsa, A, Wang, K.H, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.273 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
8G2Y
 
 | | Cryo-EM structure of ADGRF1 coupled to miniGs/q | | Descriptor: | Adhesion G-protein-coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jones, D, Rawson, S, Blacklow, S. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-10 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Tethered agonist activated ADGRF1 structure and signalling analysis reveal basis for G protein coupling.
Nat Commun, 14, 2023
|
|
1N31
 
 | | Structure of A Catalytically Inactive Mutant (K223A) of C-DES with a Substrate (Cystine) Linked to the Co-Factor | | Descriptor: | CYSTEINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
8GHV
 
 | | Cannabinoid Receptor 1-G Protein Complex | | Descriptor: | (5Z,8Z,11Z,13S,14Z)-N-[(2R)-1-hydroxypropan-2-yl]-13-methylicosa-5,8,11,14-tetraenamide, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Krishna Kumar, K, Robertson, M.J, Skiniotis, G, Kobilka, B.K. | | Deposit date: | 2023-03-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for activation of CB1 by an endocannabinoid analog.
Nat Commun, 14, 2023
|
|
7VFX
 
 | | The structure of Formyl Peptide Receptor 1 in complex with Gi and peptide agonist fMIFL | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X.K, Chen, G, Liao, Q.W, Du, Y, Hu, H.L, Ye, D.Q. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for recognition of N-formyl peptides as pathogen-associated molecular patterns.
Nat Commun, 13, 2022
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
4XAX
 
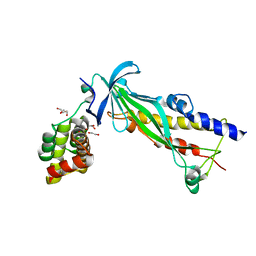 | | Crystal structure of Thermus thermophilus CarD in complex with the Thermus aquaticus RNA polymerase beta1 domain | | Descriptor: | 1,2-ETHANEDIOL, CarD, DNA-directed RNA polymerase subunit beta domain 1 | | Authors: | Chen, J, Bae, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex.
Elife, 4, 2015
|
|
5K9Y
 
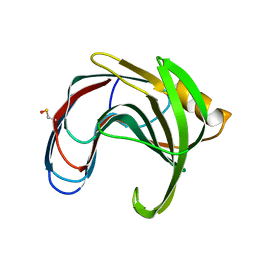 | | Crystal structure of a thermophilic xylanase A from Bacillus subtilis 1A1 quadruple mutant Q7H/G13R/S22P/S179C | | Descriptor: | Endo-1,4-beta-xylanase A | | Authors: | Pinheiro, M.P, Ferreira, T.L, Silva, S.R.B, Fuzo, C.A, Silva, S.R, Lourenzoni, M.R, Vieira, D.S, Ward, R.J, Nonato, M.C. | | Deposit date: | 2016-06-01 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The role of local residue environmental changes in thermostable mutants of the GH11 xylanase from Bacillus subtilis.
Int. J. Biol. Macromol., 97, 2017
|
|
5KCM
 
 | | Crystal structure of iron-sulfur cluster containing photolyase PhrB mutant I51W | | Descriptor: | (6-4) photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, X, Bowatte, K, Zhang, F, Lamparter, T. | | Deposit date: | 2016-06-06 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
7WB4
 
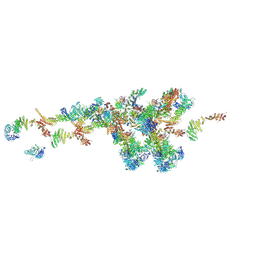 | | Cryo-EM structure of the NR subunit from X. laevis NPC | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Huang, G, Zhan, X, Shi, Y. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
4XN2
 
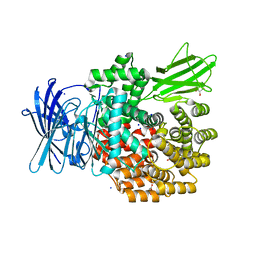 | |
4QIR
 
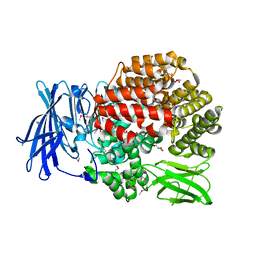 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-2-(pyridin-3-yl)ethylGlyP[CH2]Phe | | Descriptor: | 3-{[(R)-1-amino-3-(pyridin-3-yl)propyl](hydroxy)phosphoryl}-(S)-2-benzylpropanoic acid, Aminopeptidase N, GLYCEROL, ... | | Authors: | Nocek, B, Joachimiak, A, Berlicki, L, Vassiliou, S, Mucha, A. | | Deposit date: | 2014-06-01 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
1QS5
 
 | |
1QSB
 
 | |
