5J3F
 
 | | NMR solution structure of [Rp, Rp]-PT dsDNA | | Descriptor: | DNA (5'-D(*CP*GP*(RSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(RSG)P*CP*CP*G)-3') | | Authors: | Lan, W, Hu, Z, Cao, C. | | Deposit date: | 2016-03-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
1UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
5J3G
 
 | |
6F57
 
 | |
4LVL
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt+3'Thiophosphate). Mn-bound crystal structure at pH 6.8 | | Descriptor: | CHLORIDE ION, DNA (5'-D(*AP*CP*TP*TP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*AP*AP*GP*TP*AP*TP*AP*GP*TP*GP*TP*GP*(TS6))-3'), ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8XIH
 
 | | protein-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*A)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*AP*TP*TP*TP*GP*T)-3'), ... | | Authors: | Li, Z. | | Deposit date: | 2023-12-19 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | protein-DNA complex
To Be Published
|
|
7XKM
 
 | |
3O59
 
 | |
9C5Q
 
 | |
7WAZ
 
 | |
8Y0B
 
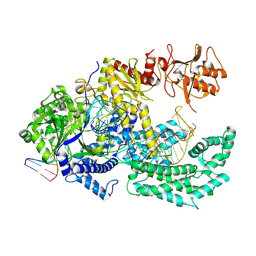 | | Crystal structure of FnCas12a in complex with pre-crRNA and 12nt target DNA | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*AP*AP*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*AP*GP*GP*AP*CP*T)-3'), DNA (5'-D(P*AP*GP*TP*CP*CP*TP*TP*TP*AP*GP*AP*TP*A)-3'), ... | | Authors: | Lin, X, Chen, J, Liu, L. | | Deposit date: | 2024-01-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA target binding-induced pre-crRNA processing in type II and V CRISPR-Cas systems.
Nucleic Acids Res., 53, 2025
|
|
8Y0C
 
 | |
8VXY
 
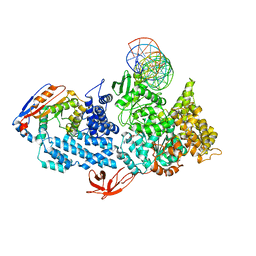 | | Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HamA, HamB, ... | | Authors: | Tuck, O.T, Hu, J.J, Doudna, J.A. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Hachiman is a genome integrity sensor.
Biorxiv, 2024
|
|
8W0A
 
 | |
6W3N
 
 | | APE1 exonuclease substrate complex D148E | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(C7R))-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6W3U
 
 | | APE1 exonuclease substrate complex R237C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6W2P
 
 | | APE1 endonuclease product complex L104R | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6W0Q
 
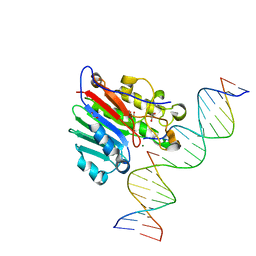 | | APE1 endonuclease product complex D148E | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-02 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6W43
 
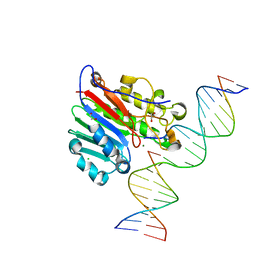 | | APE1 AP-endonuclease product complex R237C | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
8XC7
 
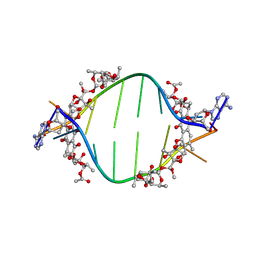 | |
3NDH
 
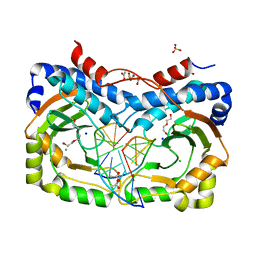 | | Restriction endonuclease in complex with substrate DNA | | Descriptor: | ACETATE ION, CHLORIDE ION, DNA (5'-D(*C*CP*AP*TP*CP*GP*CP*GP*TP*AP*C)-3'), ... | | Authors: | Firczuk, M, Wojciechowski, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | DNA intercalation without flipping in the specific ThaI-DNA complex
Nucleic Acids Res., 39, 2011
|
|
8DC2
 
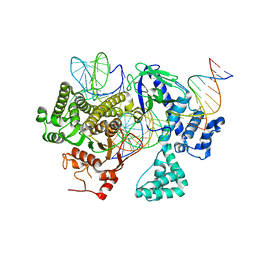 | | Cryo-EM structure of CasLambda (Cas12l) bound to crRNA and DNA | | Descriptor: | CasLambda, DNA NTS, DNA TS, ... | | Authors: | Al-Shayeb, B, Skopintsev, P, Soczek, K, Doudna, J. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors.
Cell, 185, 2022
|
|
7WB0
 
 | |
7LWA
 
 | |
7LW9
 
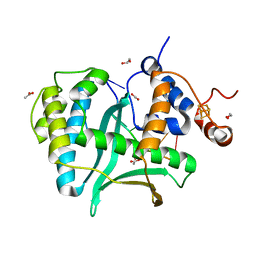 | | Human Exonuclease 5 crystal structure in complex with ssDNA, Sm, and Na | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*AP*TP*TP*GP*CP*TP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
