4NRW
 
 | | MvNei1-G86D | | Descriptor: | 5'-D(*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*C)-3', 5'-D(*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*G)-3', formamidopyrimidine-DNA glycosylase | | Authors: | Prakash, A, Doublie, S. | | Deposit date: | 2013-11-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: Activity, structure, and the effect of editing.
Dna Repair, 14, 2014
|
|
4NVB
 
 | |
4NVM
 
 | |
4NCF
 
 | |
4NWN
 
 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage T32-28 | | Descriptor: | Propanediol utilization: polyhedral bodies pduT, Uncharacterized protein | | Authors: | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
4NCX
 
 | |
4NDT
 
 | |
4NDX
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92N at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Polar cavities in proteins
To be Published
|
|
4NGN
 
 | | Crystal Structure of Glutamate Carboxypeptidase II in a complex with urea-based inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Pachl, P. | | Deposit date: | 2013-11-02 | | Release date: | 2014-06-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Rational design of urea-based glutamate carboxypeptidase II (GCPII) inhibitors as versatile tools for specific drug targeting and delivery.
Bioorg.Med.Chem., 22, 2014
|
|
4NHM
 
 | | Crystal structure of Tpa1p from Saccharomyces cerevisiae, termination and polyadenylation protein 1, in complex with N-[(1-chloro-4-hydroxyisoquinolin-3-yl)carbonyl]glycine (IOX3/UN9) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE, ... | | Authors: | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Ribosomal Oxygenase OGFOD1 Provides Insights into the Regio- and Stereoselectivity of Prolyl Hydroxylases.
Structure, 23, 2015
|
|
4MUV
 
 | | M. loti cyclic-nucleotide binding domain mutant displaying inverted ligand selectivity, cyclic-GMP bound | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated potassium channel mll3241, SODIUM ION | | Authors: | Fonseca, F, Pessoa, J, Morais-Cabral, J.H. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Determinants of ligand selectivity in a cyclic nucleotide-regulated potassium channel.
J.Gen.Physiol., 144, 2014
|
|
4MW0
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea (Chem 1392) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MT6
 
 | |
4N23
 
 | | Crystal structure of the GP2 Core Domain from the California Academy of Science Virus, monoclinic symmetry | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GP2 Ectodomain | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Dai, Z, Toro, R, Lai, J.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Glycoprotein GP2 Core Domain from the CAS Virus, a Novel Arenavirus-Like Species.
J.Mol.Biol., 426, 2014
|
|
4MWS
 
 | |
4MYE
 
 | | Cymosema roseum seed lectin structure complexed with X-man | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Cymbosema roseum mannose-specific lectin, ... | | Authors: | Barroso-Neto, I.L, Rocha, B.A.M, Teixeira, C.S, Santiago, M.Q, Souza, L.A.G, Pires, A.F, Assreuy, A.M.S, Delatorre, P, Cavada, B.S. | | Deposit date: | 2013-09-27 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cymosema roseum seed lectin structure complexed with X-man
To be Published
|
|
4MYP
 
 | | Structure of the central NEAT domain, N2, of the listerial Hbp2 protein complexed with heme | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Malmirchegini, G.R, Sawaya, M.R, Clubb, R.T. | | Deposit date: | 2013-09-27 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Mechanism of Hemin Capture by Hbp2, the Hemoglobin-binding Hemophore from Listeria monocytogenes.
J.Biol.Chem., 289, 2014
|
|
4N8C
 
 | | Three-dimensional structure of the extracellular domain of Matrix protein 2 of influenza A virus | | Descriptor: | Extracellular domain of influenza Matrix protein 2, Heavy chain of monoclonal antibody, Light chain of monoclonal antibody | | Authors: | Cho, K.J, Seok, J.H, Kim, S, Roose, K, Schepens, B, Fiers, W, Saelens, X, Kim, K.H. | | Deposit date: | 2013-10-17 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the extracellular domain of matrix protein 2 of influenza A virus in complex with a protective monoclonal antibody
J.Virol., 89, 2015
|
|
4N0E
 
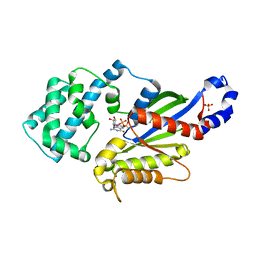 | | Crystal structure of the K345L variant of the Gi alpha1 subunit bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, SULFATE ION | | Authors: | Thaker, T.M, Preininger, A.M, Sarwar, M, Hamm, H.E, Iverson, T.M. | | Deposit date: | 2013-10-01 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Transient Interaction between the Phosphate Binding Loop and Switch I Contributes to the Allosteric Network between Receptor and Nucleotide in G alpha i1.
J.Biol.Chem., 289, 2014
|
|
4NAZ
 
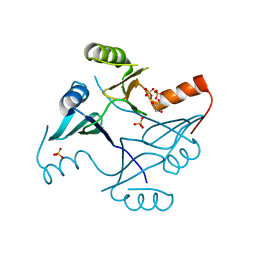 | | Crystal Structure of FosB from Staphylococcus aureus with Zn and Sulfate at 1.15 Angstrom Resolution | | Descriptor: | GLYCEROL, Metallothiol transferase FosB, SULFATE ION, ... | | Authors: | Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
4N34
 
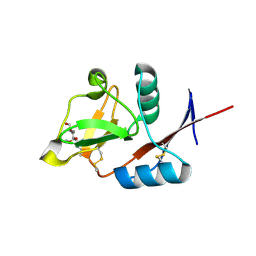 | | Structure of langerin CRD I313 with alpha-MeGlcNAc | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, methyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
4N67
 
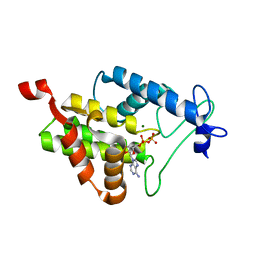 | |
4N6Q
 
 | | Crystal structure of VosA velvet domain | | Descriptor: | IODIDE ION, NITRATE ION, VosA | | Authors: | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
4NK8
 
 | |
4NNH
 
 | |
