5IO3
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - I422 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
5VFA
 
 | | RitR Mutant - C128D | | Descriptor: | Response regulator | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2017-04-07 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | RitR is an archetype for a novel family of redox sensors in the streptococci that has evolved from two-component response regulators and is required for pneumococcal colonization.
PLoS Pathog., 14, 2018
|
|
5VFP
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5IOK
 
 | |
1SHW
 
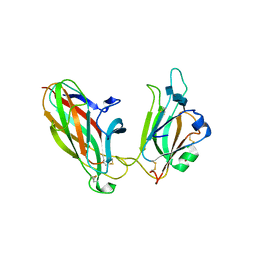 | | EphB2 / EphrinA5 Complex Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 2, Ephrin-A5, ... | | Authors: | Himanen, J.P, Chumley, M.J, Lackmann, M, Li, C, Barton, W.A, Jeffrey, P.D, Vearing, C, Geleick, D, Feldheim, D.A, Boyd, A.W. | | Deposit date: | 2004-02-26 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling
Nat.Neurosci., 7, 2004
|
|
5IOO
 
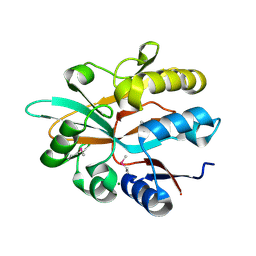 | | Accommodation of massive sequence variation in Nanoarchaeota by the C-type lectin fold | | Descriptor: | AvpA, MAGNESIUM ION | | Authors: | Handa, S, Paul, B, Miller, J, Valentine, D, Ghosh, P. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Conservation of the C-type lectin fold for accommodating massive sequence variation in archaeal diversity-generating retroelements.
Bmc Struct.Biol., 16, 2016
|
|
7LP8
 
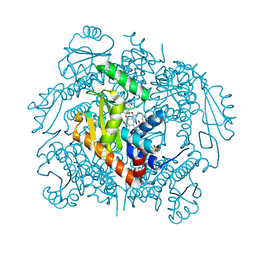 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476 | | Descriptor: | (phenylmethyl) (2~{S})-2-[5-oxidanyl-6-oxidanylidene-4-(2-pyridin-4-ylethylcarbamoyl)-1~{H}-pyrimidin-2-yl]pyrrolidine-1-carboxylate, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476
To Be Published
|
|
7LPY
 
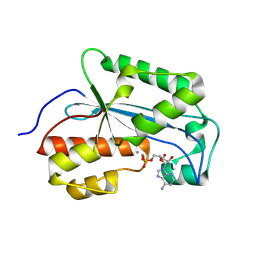 | |
5IOV
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/glycine/Ro25-6981-bound conformation | | Descriptor: | 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5VNU
 
 | | Nonheme Iron Replacement in a Biosynthetic Nitric Oxide Reductase Model Performing O2 Reduction to Water: Mn-bound FeBMb | | Descriptor: | MANGANESE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Reed, J, Shi, Y, Zhu, Q, Chakraborty, S, Mirs, E.N, Petrik, I.D, Bhagi-Damodaran, A, Ross, M, Moenne-Loccoz, P, Zhang, Y, Lu, Y. | | Deposit date: | 2017-05-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Manganese and Cobalt in the Nonheme-Metal-Binding Site of a Biosynthetic Model of Heme-Copper Oxidase Superfamily Confer Oxidase Activity through Redox-Inactive Mechanism.
J. Am. Chem. Soc., 139, 2017
|
|
7M24
 
 | |
5VNY
 
 | |
5IPV
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 1 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, McHaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.25 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5VX2
 
 | | Mcl-1 in complex with Bim-h3Pc-RT | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog,Induced myeloid leukemia cell differentiation protein Mcl-1 chimera | | Authors: | Cowan, A.D, Brouwer, J.M, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
7LPZ
 
 | |
5IQA
 
 | |
5IQO
 
 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitutions Q134E and S138E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_T4R-T6R-D13N | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Giese, C, Eras, J, Kern, A, Capitani, G, Glockshuber, R. | | Deposit date: | 2016-03-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5VIF
 
 | | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase | | Descriptor: | 2-{[(2E)-4-chlorobut-2-enoyl]amino}-2-deoxy-beta-D-glucopyranose, CKII, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, ... | | Authors: | Jiang, J, Li, B, Hu, C.-W, Worth, M, Fan, D, Li, H. | | Deposit date: | 2017-04-15 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase.
Nat. Chem. Biol., 13, 2017
|
|
7LI1
 
 | | Crystal structure of holo Moraxella catarrhalis ferric binding protein A in an open conformation | | Descriptor: | CARBONATE ION, FE (III) ION, Fe(3+) ABC transporter substrate-binding protein | | Authors: | Chan, C, Ng, D, Fraser, M.E, Schryvers, A.B. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional insights into iron acquisition from lactoferrin and transferrin in Gram-negative bacterial pathogens.
Biometals, 2022
|
|
5IR3
 
 | |
7LRR
 
 | |
5IMJ
 
 | |
7LSL
 
 | |
5IMQ
 
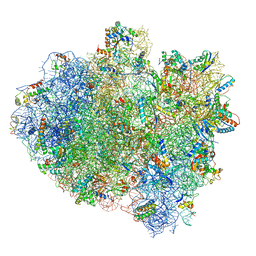 | | Structure of ribosome bound to cofactor at 3.8 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
5IRO
 
 | |
