5R88
 
 | |
5R8P
 
 | |
5R96
 
 | |
5R9O
 
 | |
5RA0
 
 | |
5MUM
 
 | | Glycoside Hydrolase BACINT_00347 | | Descriptor: | 1,2-ETHANEDIOL, BACINT_00347, PENTAETHYLENE GLYCOL | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UYO
 
 | | Solution NMR structure of the de novo mini protein HEEH_rd4_0097 | | Descriptor: | HEEH_rd4_0097 | | Authors: | Lemak, A, Rocklin, G.J, Houliston, S, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UP5
 
 | | Solution structure of the de novo mini protein EHEE_rd1_0284 | | Descriptor: | EHEE_rd1_0284 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UP1
 
 | | Solution structure of the de novo mini protein EEHEE_rd3_1049 | | Descriptor: | EEHEE_rd3_1049 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UOI
 
 | | Solution structure of the de novo mini protein HHH_rd1_0142 | | Descriptor: | HHH_rd1_0142 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5C7L
 
 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase apo form | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
4A60
 
 | | Crystal structure of human testis-specific fatty acid binding protein 9 (FABP9) | | Descriptor: | 1,2-ETHANEDIOL, FATTY ACID-BINDING PROTEIN 9 TESTIS LIPID-BINDING PROTEIN, TLBP, ... | | Authors: | Muniz, J.R.C, Kiyani, W, Shrestha, L, Froese, D.S, Krojer, T, Vollmar, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, von Delft, F, Yue, W.W. | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Crystal Structure of the Human Fabp9A
To be Published
|
|
4FLP
 
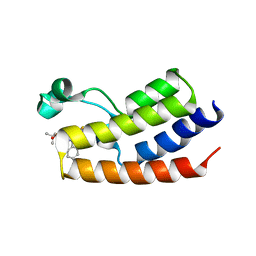 | | Crystal Structure of the first bromodomain of human BRDT in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Bromodomain testis-specific protein, POTASSIUM ION | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Canning, P, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bradner, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-15 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Small-Molecule Inhibition of BRDT for Male Contraception.
Cell(Cambridge,Mass.), 150, 2012
|
|
5C7O
 
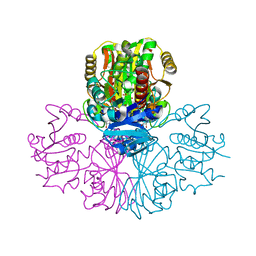 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase holo form with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
4UFC
 
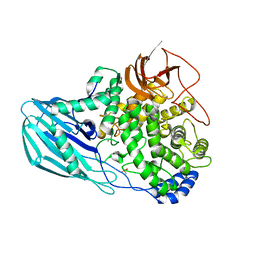 | | Crystal structure of the GH95 enzyme BACOVA_03438 | | Descriptor: | CACODYLATE ION, CALCIUM ION, GH95, ... | | Authors: | Rogowski, A, Briggs, J.A, Mortimer, J.C, Tryfona, T, Terrapon, N, Lowe, E.C, Basle, A, Morland, C, Day, A.M, Zheng, H, Rogers, T.E, Thompson, P, Hawkins, A.R, Yadav, M.P, Henrissat, B, Martens, E.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Glycan Complexity Dictates Microbial Resource Allocation in the Large Intestine.
Nat.Commun., 6, 2015
|
|
5ABW
 
 | | Neutrophil elastase inhibitors for the treatment of (cardio)pulmonary diseases | | Descriptor: | 1-(3-CHLOROPHENYL)-5-(3,5-DIMETHYLISOXAZOL-4-YL)-6-METHYL-N-[4-(METHYLSULFONYL)BENZYL]-2-OXO-1,2-DIHYDROPYRIDINE-3-CARBOXAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | von Nussbaum, F, Li, V.M, Schaefer, M. | | Deposit date: | 2015-08-10 | | Release date: | 2015-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Neutrophil elastase inhibitors for the treatment of (cardio)pulmonary diseases: Into clinical testing with pre-adaptive pharmacophores.
Bioorg. Med. Chem. Lett., 25, 2015
|
|
2FBM
 
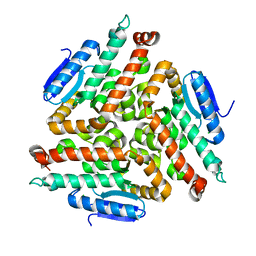 | | Acetyltransferase domain of CDY1 | | Descriptor: | CHLORIDE ION, Y chromosome chromodomain protein 1, telomeric isoform b | | Authors: | Min, J.R, Antoshenko, T, Hong, W, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-09 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of Acetyltransferases domain of Human Testis-specific chromodomain protein Y 1
To be Published
|
|
4KCX
 
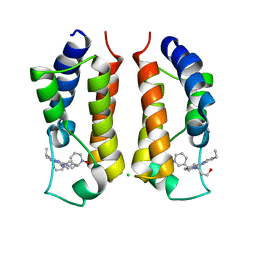 | | BRDT in complex with Dinaciclib | | Descriptor: | 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, Bromodomain testis-specific protein, CHLORIDE ION | | Authors: | Martin, M.P, Schonbrunn, E. | | Deposit date: | 2013-04-24 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclin-dependent kinase inhibitor dinaciclib interacts with the acetyl-lysine recognition site of bromodomains.
Acs Chem.Biol., 8, 2013
|
|
4CJX
 
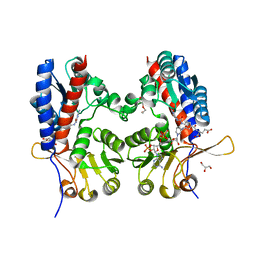 | | The crystal structure of Trypanosoma brucei N5, N10- methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) complexed with NADP cofactor and inhibitor | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, C-1-TETRAHYDROFOLATE SYNTHASE, CYTOPLASMIC, ... | | Authors: | Eadsforth, T.C, Hunter, W.N. | | Deposit date: | 2013-12-23 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of 2,4-Diamino-6-Oxo-1,6-Dihydropyrimidin-5-Yl Ureido Based Inhibitors of Trypanosoma Brucei Fold and Testing for Antiparasitic Activity.
J.Med.Chem., 58, 2015
|
|
3MS9
 
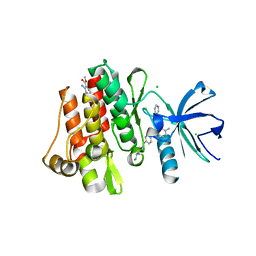 | | ABL kinase in complex with imatinib and a fragment (FRAG1) in the myristate pocket | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, Tyrosine-protein kinase ABL1, ... | | Authors: | Cowan-Jacob, S.W, Rummel, G, Fendrich, G. | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding or bending: distinction of allosteric Abl kinase agonists from antagonists by an NMR-based conformational assay.
J.Am.Chem.Soc., 132, 2010
|
|
8OTZ
 
 | | 48-nm repeat of the native axonemal doublet microtubule from bovine sperm | | Descriptor: | ATP6V1F neighbor, CFAP97 domain containing 1, Chromosome 13 C20orf85 homolog, ... | | Authors: | Leung, M.R, Zeng, J, Zhang, R, Zeev-Ben-Mordehai, T. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural specializations of the sperm tail.
Cell, 186, 2023
|
|
8BJ2
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in the closed state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, SODIUM ION, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
8BJ3
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in complex with histidinol-phosphate | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, [(2~{S})-3-(1~{H}-imidazol-4-yl)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]propyl] dihydrogen phosphate, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
2YHD
 
 | | Human androgen receptor in complex with AF2 small molecule inhibitor | | Descriptor: | 4-(2,3-dihydro-1H-perimidin-2-yl)benzene-1,2-diol, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | AxerioCilies, P, Lack, N.A, ShashiNayana, M.R, Chan, K.H, Yeung, A, LeBlanc, E, Guns, E, Rennie, P, Cherkasov, A. | | Deposit date: | 2011-04-28 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Androgen Receptor Activation Function-2 (Af2) Site Identified Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
8E3Z
 
 | | Cryo-EM structure of the VPAC1R-VIP-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Piper, S.J, Danev, R, Sexton, P, Wootten, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Understanding VPAC receptor family peptide binding and selectivity.
Nat Commun, 13, 2022
|
|
