2YK1
 
 | | Structure of human anti-nicotine Fab fragment in complex with nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, FAB FRAGMENT, HEAVY CHAIN, ... | | Authors: | Tars, K, Kotelovica, S, Lipowsky, G, Bauer, M, Beerli, R, Bachmann, M, Maurer, P. | | Deposit date: | 2011-05-25 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Different Binding Modes of Free and Carrier-Protein-Coupled Nicotine in a Human Monoclonal Antibody.
J.Mol.Biol., 415, 2012
|
|
7I57
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0036798-001 (A71EV2A-x4794) | | Descriptor: | 1-[(3S)-3-hydroxypiperidin-1-yl]-2-[2-(methylsulfanyl)phenyl]ethan-1-one, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I4V
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0036758-001 (A71EV2A-x4746) | | Descriptor: | 1-[(3S)-3-(difluoromethyl)piperidin-1-yl]-2-[2-(methylsulfanyl)phenyl]ethan-1-one, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I4X
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0036773-001 (A71EV2A-x4751) | | Descriptor: | 1-[(3S)-3-(fluoromethyl)piperidin-1-yl]-2-[2-(methylsulfanyl)phenyl]ethan-1-one, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.147 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
4ZG7
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-({6-chloro-7-fluoro-2-methyl-1-[2-oxo-2-(spiro[cyclopropane-1,3'-indol]-1'(2'H)-yl)ethyl]-1H-indol-3-yl}sulfanyl)-2-fluorobenzoic acid, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
5OHS
 
 | | A GH31 family sulfoquinovosidase mutant D455N in complex with pNPSQ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 4-nitrophenyl alpha-D-6-sulfoquinovoside, ... | | Authors: | Jin, Y, Williams, S.J, Goddard-Borger, E, Davies, G.J. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Biochemical Insights into the Function and Evolution of Sulfoquinovosidases.
ACS Cent Sci, 4, 2018
|
|
8CGN
 
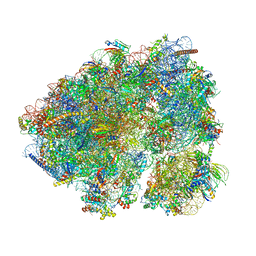 | | Non-rotated 80S S. cerevisiae ribosome with ligands | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
2VZQ
 
 | |
3G9E
 
 | | Aleglitaar. a new. potent, and balanced dual ppara/g agonist for the treatment of type II diabetes | | Descriptor: | (2S)-2-methoxy-3-{4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Ruf, A, Benz, J, Bernardeau, A, Binggeli, A, Blum, D, Boehringer, M, Grether, U, Hilpert, H, Kuhn, B, Maerki, H.P, Meyer, M, Puenterner, K, Raab, S, Schlatter, D, Gsell, B, Stihle, M, Mohr, P. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aleglitazar, a new, potent, and balanced dual PPARalpha/gamma agonist for the treatment of type II diabetes.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4CGI
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | 5-[[(1S,2S)-2-(6-azanylhexanoylamino)-2,3-dihydro-1H-inden-1-yl]methyl]-1,3-benzodioxole-4-carboxylic acid, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-25 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
2W12
 
 | | High-resolution crystal structure of the P-I snake venom metalloproteinase BaP1 in complex with a peptidomimetic: insights into inhibitor binding | | Descriptor: | (2R,3R)-N^1^-[(1S)-2,2-DIMETHYL-1-(METHYLCARBAMOYL)PROPYL]-N^4^-HYDROXY-2-(2-METHYLPROPYL)-3-{[(1,3-THIAZOL-2-YLCARBONYL)AMINO]METHYL}BUTANEDIAMIDE, GLYCEROL, ZINC ION, ... | | Authors: | Lingott, T.J, Schleberger, C, Gutierrez, J.M, Merfort, I. | | Deposit date: | 2008-10-14 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | High-Resolution Crystal Structure of the Snake Venom Metalloproteinase Bap1 Complexed with a Peptidomimetic: Insight Into Inhibitor Binding.
Biochemistry, 48, 2009
|
|
2W13
 
 | | High-resolution crystal structure of the P-I snake venom metalloproteinase BaP1 in complex with a peptidomimetic: insights into inhibitor binding | | Descriptor: | (2R,3R)-N^1^-[(1S)-2,2-DIMETHYL-1-(METHYLCARBAMOYL)PROPYL]-N^4^-HYDROXY-2-(2-METHYLPROPYL)-3-{[(1,3-THIAZOL-2-YLCARBONYL)AMINO]METHYL}BUTANEDIAMIDE, ACETATE ION, GLYCEROL, ... | | Authors: | Lingott, T.J, Schleberger, C, Gutierrez, J.M, Merfort, I. | | Deposit date: | 2008-10-14 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | High-Resolution Crystal Structure of the Snake Venom Metalloproteinase Bap1 Complexed with a Peptidomimetic: Insight Into Inhibitor Binding.
Biochemistry, 48, 2009
|
|
1H03
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
6EDS
 
 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Glucagon and Substrate-selective Macrocyclic Inhibitor 63 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, G.A, Seeliger, M.A, Maianti, J.P, Liu, D.R, Welsh, A.J. | | Deposit date: | 2018-08-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.18071723 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
1UPR
 
 | | Crystal structure of the PEPP1 pleckstrin homology domain in complex with Inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, PLECKSTRIN HOMOLOGY DOMAIN-CONTAINING FAMILY A MEMBER 4 | | Authors: | Milburn, C.C, Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Pleckstrin Homology Domain of Pepp1
To be Published
|
|
3PGP
 
 | | Crystal structure of PA4794 - GNAT superfamily protein in complex with AcCoA | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
2ANL
 
 | | X-ray crystal structure of the aspartic protease plasmepsin 4 from the malarial parasite plasmodium malariae bound to an allophenylnorstatine based inhibitor | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, plasmepsin IV | | Authors: | Clemente, J.C, Govindasamy, L, Madabushi, A, Fisher, S.Z, Moose, R.E, Yowell, C.A, Hidaka, K, Kimura, T, Hayashi, Y, Kiso, Y, Agbandje-McKenna, M, Dame, J.B, Dunn, B.M, McKenna, R. | | Deposit date: | 2005-08-11 | | Release date: | 2006-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the aspartic protease plasmepsin 4 from the malarial parasite Plasmodium malariae bound to an allophenylnorstatine-based inhibitor.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5O43
 
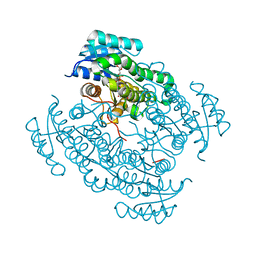 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with a non-steroidal 2,6-pyridinketone inhibitor. | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, 2-fluoranyl-3-[6-[(4-fluoranyl-3-oxidanyl-phenyl)-methyl-amino]pyridin-2-yl]phenol, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Heine, A, Braun, F, Klebe, G, Marchais-Oberwinkler, S. | | Deposit date: | 2017-05-25 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design and profiling of novel 17 beta-HSD14 inhibitors.
Eur J Med Chem, 155, 2018
|
|
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
2VTH
 
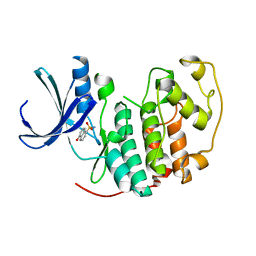 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | Descriptor: | 5-hydroxynaphthalene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2AOA
 
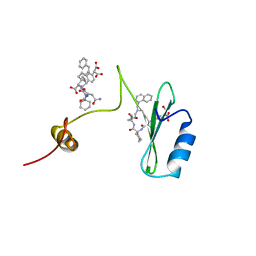 | | Crystal structures of a high-affinity macrocyclic peptide mimetic in complex with the Grb2 SH2 domain | | Descriptor: | 2-(4-((9S,10S,14S,Z)-18-(2-AMINO-2-OXOETHYL)-9-(CARBOXYMETHYL)-14-(NAPHTHALEN-1-YLMETHYL)-8,17,20-TRIOXO-7,16,19-TRIAZASPIRO[5.14]ICOS-11-EN-10-YL)PHENYL)MALONIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Growth factor receptor-bound protein 2 | | Authors: | Phan, J, Shi, Z.D, Burke, T.R, Waugh, D.S. | | Deposit date: | 2005-08-12 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structures of a High-affinity Macrocyclic Peptide Mimetic in Complex with the Grb2 SH2 Domain.
J.Mol.Biol., 353, 2005
|
|
5SYT
 
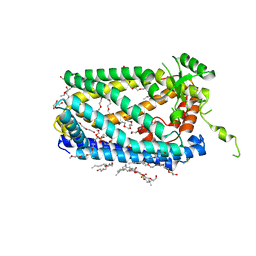 | | Crystal Structure of ZMPSTE24 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX prenyl protease 1 homolog, ... | | Authors: | Clark, K, Jenkins, J.L, Fedoriw, N, Dumont, M.E, Membrane Protein Structural Biology Consortium (MPSBC) | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human CaaX protease ZMPSTE24 expressed in yeast: Structure and inhibition by HIV protease inhibitors.
Protein Sci., 26, 2017
|
|
4QVV
 
 | | yCP beta5-A49V mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
3B4V
 
 | | X-Ray structure of Activin in complex with FSTL3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Inhibin beta A chain, ... | | Authors: | Thompson, T.B. | | Deposit date: | 2007-10-24 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The structure of FSTL3.activin A complex. Differential binding of N-terminal domains influences follistatin-type antagonist specificity.
J.Biol.Chem., 283, 2008
|
|
6B0V
 
 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
