3CII
 
 | | Structure of NKG2A/CD94 bound to HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen peptide, HLA class I histocompatibility antigen, ... | | Authors: | Strong, R.K, Kaiser, B.K, Pizarro, J.C. | | Deposit date: | 2008-03-11 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.41 Å) | | Cite: | Structural basis for NKG2A/CD94 recognition of HLA-E.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1K4R
 
 | | Structure of Dengue Virus | | Descriptor: | MAJOR ENVELOPE PROTEIN E | | Authors: | Kuhn, R.J, Zhang, W, Rossmann, M.G, Pletnev, S.V, Corver, J, Lenches, E, Jones, C.T, Mukhopadhyay, S, Chipman, P.R, Strauss, E.G, Baker, T.S, Strauss, J.H. | | Deposit date: | 2001-10-08 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Structure of dengue virus: implications for flavivirus organization, maturation, and fusion.
Cell(Cambridge,Mass.), 108, 2002
|
|
2C0B
 
 | | Catalytic domain of E. coli RNase E in complex with 13-mer RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP*UP*G)-3', MAGNESIUM ION, RIBONUCLEASE E, ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-08-30 | | Release date: | 2005-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
2C4R
 
 | | Catalytic domain of E. coli RNase E | | Descriptor: | MAGNESIUM ION, RIBONUCLEASE E, SSRNA MOLECULE: 5'-R(*AP*CP*AP*GP*UP*AP*UP*UP*UP*GP)-3', ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Luisi, B.F. | | Deposit date: | 2005-10-21 | | Release date: | 2005-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
2BX2
 
 | | Catalytic domain of E. coli RNase E | | Descriptor: | MAGNESIUM ION, RIBONUCLEASE E, RNA (5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP* UP*GP*UP*U)-3'), ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
2BZ2
 
 | | Solution structure of NELF E RRM | | Descriptor: | NEGATIVE ELONGATION FACTOR E | | Authors: | Schweimer, K, Rao, J.N, Neumann, L, Rosch, P, Wohrl, B.M. | | Deposit date: | 2005-08-10 | | Release date: | 2006-08-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the RNA-recognition motif of NELF E, a cellular negative transcription elongation factor involved in the regulation of HIV transcription.
Biochem. J., 400, 2006
|
|
2R6P
 
 | | Fit of E protein and Fab 1A1D-2 into 24 angstrom resolution cryoEM map of Fab complexed with dengue 2 virus. | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1T4R
 
 | | arginase-descarboxy-nor-NOHA complex | | Descriptor: | 3-{[(E)-AMINO(HYDROXYIMINO)METHYL]AMINO}PROPAN-1-AMINIUM, Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Shoufa, H, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
7RQ8
 
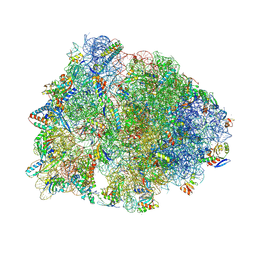 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
7RQ9
 
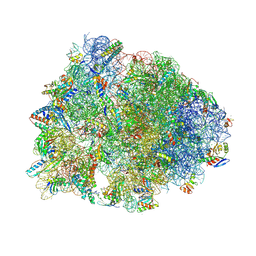 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
2R99
 
 | | Crystal structure of cyclophilin ABH-like domain of human peptidylprolyl isomerase E isoform 1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-12 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
9B0T
 
 | | Cryo-EM structure of E227Q variant of uMtCK1 in complex with transition state analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-12 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
9B14
 
 | | Cryo-EM structure of human uMtCK1 in complex with transition state analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-13 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
9B0U
 
 | | Cryo-EM structure of E227Q variant of uMtCK1 incubated with ADP and phosphocreatine at pH 8.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-12 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
7RSV
 
 | | Structure of the VPS34 kinase domain with compound 5 | | Descriptor: | (5aS,8aR,9S)-2-[(3R)-3-methylmorpholin-4-yl]-5,5a,6,7,8,8a-hexahydro-4H-cyclopenta[e]pyrazolo[1,5-a]pyrazin-4-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
6X3S
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with bicuculline methbromide | | Descriptor: | (5S)-6,6-dimethyl-5-[(6R)-8-oxo-6,8-dihydrofuro[3,4-e][1,3]benzodioxol-6-yl]-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-6-ium, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, J.J, Gharpure, A, Teng, J, Zhuang, Y, Howard, R.J, Zhu, S, Noviello, C.M, Walsh, R.M, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Shared structural mechanisms of general anaesthetics and benzodiazepines.
Nature, 585, 2020
|
|
1SS3
 
 | | Solution structure of Ole e 6, an allergen from olive tree pollen | | Descriptor: | Pollen allergen Ole e 6 | | Authors: | Trevino, M.A, Garcia-Mayoral, M.F, Barral, P, Villalba, M, Santoro, J, Rico, M, Rodriguez, R, Bruix, M. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Ole e 6, a Major Allergen from Olive Tree Pollen.
J.Biol.Chem., 279, 2004
|
|
1T3A
 
 | | Crystal structure of Clostridium botulinum neurotoxin type E catalytic domain | | Descriptor: | CHLORIDE ION, ZINC ION, neurotoxin type E | | Authors: | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | Deposit date: | 2004-04-26 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
5QCN
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-5-[(3~{S})-3-ethoxycarbonylpiperidin-1-yl]carbonyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-5-[(3~{S})-3-ethoxycarbonylpiperidin-1-yl]carbonyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
5QCL
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
6I1S
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with FKBP12 and the inhibitor E6201 | | Descriptor: | (4~{S},5~{R},6~{Z},9~{S},10~{S},12~{E})-16-(ethylamino)-4,5-dimethyl-9,10,18-tris(oxidanyl)-3-oxabicyclo[12.4.0]octadeca-1(14),6,12,15,17-pentaene-2,8-dione, 1,2-ETHANEDIOL, Activin receptor type-1, ... | | Authors: | Williams, E.P, Pinkas, D.M, Fortin, J, Newman, J.A, Bradshaw, W.J, Mahajan, P, Kupinska, K, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-10-30 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mutant ACVR1 Arrests Glial Cell Differentiation to Drive Tumorigenesis in Pediatric Gliomas.
Cancer Cell, 37, 2020
|
|
1SU1
 
 | | Structural and biochemical characterization of Yfce, a phosphoesterase from E. coli | | Descriptor: | Hypothetical protein yfcE, SULFATE ION, ZINC ION | | Authors: | Miller, D.J, Shuvalova, L, Evdokimova, E, Savchenko, A, Yakunin, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biochemical characterization of a novel Mn2+-dependent phosphodiesterase encoded by the yfcE gene.
Protein Sci., 16, 2007
|
|
6I2K
 
 | | Structure of EV71 complexed with its receptor SCARB2 | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Zhao, Y, Kotecha, A, Fry, E.E, Kelly, J, Wang, X, Rao, Z, Rowlands, D.J, Ren, J, Stuart, D.I. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2.
Nat Microbiol, 4, 2019
|
|
6X0C
 
 | | Tryptophan Synthase mutant beta-Q114A in complex with Cesium ion at the metal coordination site and aminoacrylate and benzimidazole at the enzyme beta site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BENZIMIDAZOLE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2020-05-15 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | External aldimine form of Tryptophan Synthase mutant beta-Q114A in complex with Cesium ion at the metal coordination site and benzimidazole at the enzyme beta site.
To be Published
|
|
4BWL
 
 | | Structure of the Y137A mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate, N-acetyl-D-mannosamine and N- acetylneuraminic acid | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, 5-(acetylamino)-3,5-dideoxy-D-glycero-D-galacto-non-2-ulosonic acid, N-ACETYLNEURAMINATE LYASE, ... | | Authors: | Campeotto, I, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2013-07-03 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Reaction Mechanism of N-Acetylneuraminic Acid Lyase Revealed by a Combination of Crystallography, Qm/Mm Simulation and Mutagenesis.
Acs Chem.Biol., 9, 2014
|
|
