3VT4
 
 | | Crystal structures of rat VDR-LBD with R270L mutation | | Descriptor: | (1R,2Z,3R,5E,7E)-17-{(1S)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethylidene)-9,10-secoestra-5,7,16-triene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
3EYC
 
 | | New crystal structure of human tear lipocalin in complex with 1,4-butanediol in space group P21 | | Descriptor: | 1,4-BUTANEDIOL, Lipocalin-1 | | Authors: | Breustedt, D.A, Keil, L, Skerra, A. | | Deposit date: | 2008-10-20 | | Release date: | 2009-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new crystal form of human tear lipocalin reveals high flexibility in the loop region and induced fit in the ligand cavity
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5XPM
 
 | | Crystal structure of VDR-LBD complexed with 22S-Butyl-25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},3~{S})-3-[(3~{S})-3-(4-hydroxyphenyl)-3-methoxy-propyl]heptan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
7T7Z
 
 | | The crystal structure of family 8 carbohydrate-binding module from Dictyostelium discoideum | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, Endoglucanase, ... | | Authors: | Marcelo, M.V, Campos, B.M, Squina, F.M. | | Deposit date: | 2021-12-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Unique properties of a Dictyostelium discoideum carbohydrate-binding module expand our understanding of CBM-ligand interactions.
J.Biol.Chem., 298, 2022
|
|
7T7Y
 
 | |
5XPL
 
 | | Crystal structure of VDR-LBD complexed with 22S-butyl-25-hydroxyphenyl-2-methylidene-19,26,27-trinor-25-oxo-1-hydroxyvitamin D3 | | Descriptor: | (4~{S})-4-[(1~{R})-1-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl )cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]ethyl]-1-(4-hydroxyphenyl)octan-1-one, Nuclear receptor coactivator 2, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
3AXA
 
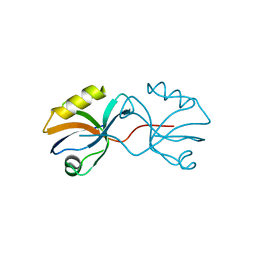 | | Crystal structure of afadin PDZ domain in complex with the C-terminal peptide from nectin-3 | | Descriptor: | Afadin, Nectin-3 | | Authors: | Fujiwara, Y, Goda, N, Narita, H, Satomura, K, Nakagawa, A, Sakisaka, T, Suzuki, M, Hiroaki, H. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of afadin PDZ domain-nectin-3 complex shows the structural plasticity of the ligand-binding site.
Protein Sci., 24, 2015
|
|
8GCK
 
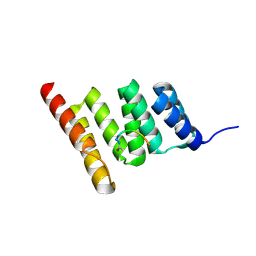 | | Crystal structure of the human CHIP-TPR domain in complex with a 6mer acetylated tau peptide | | Descriptor: | ACE-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | Authors: | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.36823535 Å) | | Cite: | Intersecting PTMs regulate clearance of pathogenic tau by the ubiquitin ligase CHIP.
To Be Published
|
|
1LN2
 
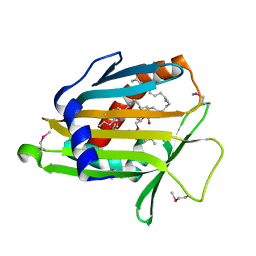 | | Crystal Structure of Human Phosphatidylcholine Transfer Protein in Complex with Dilinoleoylphosphatidylcholine (Seleno-Met Protein) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylcholine transfer protein | | Authors: | Roderick, S.L, Chan, W.W, Agate, D.S, Olsen, L.R, Vetting, M.W, Rajashankar, K.R, Cohen, D.E. | | Deposit date: | 2002-05-02 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human phosphatidylcholine transfer protein in complex with its ligand.
Nat.Struct.Biol., 9, 2002
|
|
6LW5
 
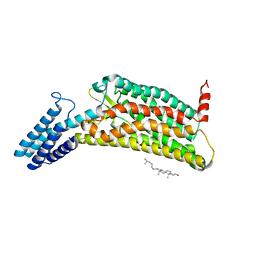 | | Crystal structure of the human formyl peptide receptor 2 in complex with WKYMVm | | Descriptor: | CHOLESTEROL, Soluble cytochrome b562,N-formyl peptide receptor 2, TRP-LYS-TYR-MET-VAL-QXV | | Authors: | Chen, T, Zong, X, Zhang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2020-02-07 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Nat Commun, 11, 2020
|
|
3C9A
 
 | | High Resolution Crystal Structure of Argos bound to the EGF domain of Spitz | | Descriptor: | BROMIDE ION, Protein giant-lens, Protein spitz | | Authors: | Klein, D.E, Stayrook, S.E, Shi, F, Narayan, K, Lemmon, M.A. | | Deposit date: | 2008-02-15 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for EGFR ligand sequestration by Argos.
Nature, 453, 2008
|
|
5QCQ
 
 | | Crystal structure of BACE complex with BMC025 | | Descriptor: | (2R,4S,5S)-N-butyl-4-hydroxy-2,7-dimethyl-5-{[N-(4-methylpentanoyl)-L-methionyl]amino}octanamide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1GSA
 
 | | STRUCTURE OF GLUTATHIONE SYNTHETASE COMPLEXED WITH ADP AND GLUTATHIONE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTATHIONE, GLUTATHIONE SYNTHETASE, ... | | Authors: | Hara, T, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-06-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A pseudo-michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 A resolution.
Biochemistry, 35, 1996
|
|
6ZW0
 
 | |
6ZVZ
 
 | |
5QD5
 
 | | Crystal structure of BACE complex with BMC009 | | Descriptor: | (10S,12S)-17-chloro-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-10-methyl-7-oxa-2,13,18-triazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
4ANK
 
 | | Crystallographic study of novel transthyretin ligands exhibiting negative-cooperativity between two T4 binding sites. | | Descriptor: | TRANSTHYRETIN | | Authors: | Tomar, D, Khan, T, Singh, R.R, Mishra, S, Gupta, S, Surolia, A, Salunke, D.M. | | Deposit date: | 2012-03-20 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Study of Novel Transthyretin Ligands Exhibiting Negative-Cooperativity between Two Thyroxine Binding Sites.
Plos One, 7, 2012
|
|
7P3A
 
 | | N-terminal domain of CGI-99 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Kroupova, A, Jinek, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular architecture of the human tRNA ligase complex.
Elife, 10, 2021
|
|
3CDS
 
 | |
7B7N
 
 | | Human herpesvirus-8 gH/gL in complex with EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Pederzoli, R, Guardado-Calvo, P, Rey, F.A, Backovic, M. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Human herpesvirus 8 molecular mimicry of ephrin ligands facilitates cell entry and triggers EphA2 signaling.
Plos Biol., 19, 2021
|
|
1R9Q
 
 | | structure analysis of ProX in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding periplasmic protein, UNKNOWN ATOM OR ION | | Authors: | Schiefner, A, Breed, J, Bosser, L, Kneip, S, Gade, J, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cation-pi Interactions as Determinants for Binding of the Compatible Solutes Glycine Betaine and Proline Betaine by the Periplasmic Ligand-binding Protein ProX from Escherichia coli
J.BIOL.CHEM., 279, 2004
|
|
4AOP
 
 | |
8FEG
 
 | | CryoEM structure of Kappa Opioid Receptor bound to a semi-peptide and Gi1 | | Descriptor: | ACE-TYR-ALA-DTY-THR-THR-CYS-THR-DPN-XT9, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-12-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Design and structural validation of peptide-drug conjugate ligands of the kappa-opioid receptor.
Nat Commun, 14, 2023
|
|
6MWZ
 
 | | LasR LBD T75V/Y93F/A127W:BB0126 | | Descriptor: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1S,3S,5S)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, ALA-HIS-HIS-HIS-HIS-ALA, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
7MRZ
 
 | | Structure of GDF11 bound to fused ActRIIB-ECD and Alk4-ECD with Anti-ActRIIB Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B,Activin receptor type-1B, Fab Heavy Chain, ... | | Authors: | Goebel, E.J, Kattamuri, C, Gipson, G.R, Thompson, T.B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
