1TJH
 
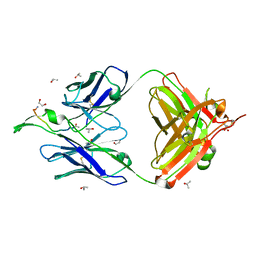 | | Crystal Structure of the broadly neutralizing anti-HIV-1 antibody 2F5 in complex with a gp41 11mer epitope | | Descriptor: | 1,2-ETHANEDIOL, Envelope glycoprotein GP41, ISOPROPYL ALCOHOL, ... | | Authors: | Ofek, G, Tang, M, Sambor, A, Katinger, H, Mascola, J.R, Wyatt, R, Kwong, P.D. | | Deposit date: | 2004-06-04 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope
J.Virol., 78, 2004
|
|
1W1A
 
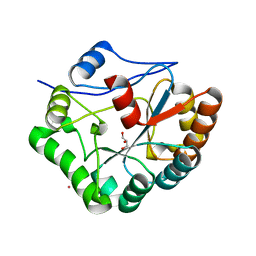 | | Structure of Bacillus subtilis PdaA in complex with NAG, a family 4 Carbohydrate esterase. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CADMIUM ION, GLYCEROL, ... | | Authors: | Blair, D.E, van Aalten, D.M.F. | | Deposit date: | 2004-06-18 | | Release date: | 2005-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of Bacillus subtilis PdaA, a family 4 carbohydrate esterase, and a complex with N-acetyl-glucosamine.
FEBS Lett., 570, 2004
|
|
1VJ9
 
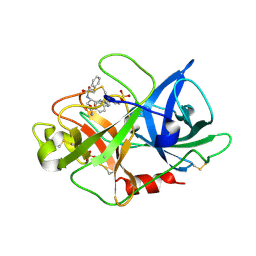 | | Urokinase Plasminogen Activator B-Chain-JT464 Complex | | Descriptor: | N-(BENZYLSULFONYL)-L-SERYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}-O-BENZYL-L-SERINAMIDE, SULFATE ION, plasminogen activator, ... | | Authors: | Schweinitz, A, Steinmetzer, T, Banke, I.J, Arlt, M.J.E, Stuerzebecher, A, Schuster, O, Geissler, A, Giersiefen, H, Zeslawska, E, Jacob, U, Kruger, A, Stuerzebecher, J. | | Deposit date: | 2004-02-03 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of novel and selective inhibitors of urokinase-type plasminogen activator with improved pharmacokinetic properties for use as antimetastatic agents
J.Biol.Chem., 279, 2004
|
|
2GAA
 
 | |
2JEW
 
 | |
2JH6
 
 | | Human Thrombin Hirugen Inhibitor complex | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHANESULFONAMIDE, CALCIUM ION, HIRUDIN IIIA, ... | | Authors: | Senger, S, Chan, C, Convery, M.A, Hubbard, J.A, Young, R.J, Shah, G.P, Watson, N.S. | | Deposit date: | 2007-02-20 | | Release date: | 2007-05-08 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Sulfonamide-Related Conformational Effects and Their Importance in Structure-Based Design.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2JJV
 
 | | Structure of human signal regulatory protein (sirp) beta(2) | | Descriptor: | CHLORIDE ION, SIGNAL-REGULATORY PROTEIN BETA 1., SULFATE ION | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Paired Receptor Specificity Explained by Structures of Signal Regulatory Proteins Alone and Complexed with Cd47.
Mol.Cell, 31, 2008
|
|
2LS0
 
 | |
2L94
 
 | | Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA_(45-MER) | | Authors: | Marcheschi, R.J, Tonelli, M, Kumar, A, Butcher, S.E. | | Deposit date: | 2011-01-29 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 Frameshift Site RNA Bound to a Small Molecule Inhibitor of Viral Replication.
Acs Chem.Biol., 6, 2011
|
|
2LPW
 
 | | human CEB25 minisatellite G-quadruplex | | Descriptor: | DNA (26-MER) | | Authors: | Amrane, S, Adrian, M, Heddi, B, Serero, A, Nicolas, A, Mergny, J.L, Phan, A.T. | | Deposit date: | 2012-02-20 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Formation of Pearl-Necklace Monomorphic G-Quadruplexes in the Human CEB25 Minisatellite.
J.Am.Chem.Soc., 134, 2012
|
|
2LR1
 
 | |
2LID
 
 | |
2LEF
 
 | | LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*CP*CP*CP*TP*TP*TP*GP*AP*AP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*TP*TP*CP*AP*AP*AP*GP*GP*GP*TP*G)-3'), PROTEIN (LYMPHOID ENHANCER-BINDING FACTOR) | | Authors: | Li, X, Love, J.J, Case, D.A, Wright, P.E. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA bending by the architectural transcription factor LEF-1.
Nature, 376, 1995
|
|
2L5A
 
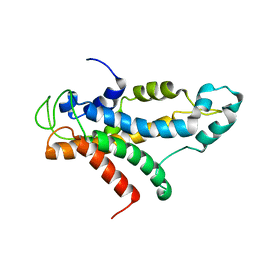 | | Structural basis for recognition of centromere specific histone H3 variant by nonhistone Scm3 | | Descriptor: | Histone H3-like centromeric protein CSE4, Protein SCM3, Histone H4 | | Authors: | Zhou, Z, Feng, H, Zhou, B, Ghirlando, R, Hu, K, Zwolak, A, Jenkins, L, Xiao, H, Tjandra, N, Wu, C, Bai, Y. | | Deposit date: | 2010-10-28 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.
Nature, 472, 2011
|
|
2LIC
 
 | |
119D
 
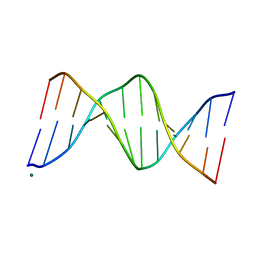 | |
144D
 
 | | MINOR GROOVE BINDING OF SN6999 TO AN ALKYLATED DNA: MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG)-SN6999 COMPLEX | | Descriptor: | 1-METHYL-4-[4-[4-(4-(1-METHYLQUINOLINIUM)AMINO)BENZAMIDO]ANILINO]PYRIDINIUM, DNA (5'-D(*CP*GP*CP*(G36)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Gao, Y.-G, Sriram, M, Denny, W.A, Wang, A.H.-J. | | Deposit date: | 1993-10-26 | | Release date: | 1995-05-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Minor groove binding of SN6999 to an alkylated DNA: molecular structure of d(CGC[e6G]AATTCGCG)-SN6999 complex.
Biochemistry, 32, 1993
|
|
1A6H
 
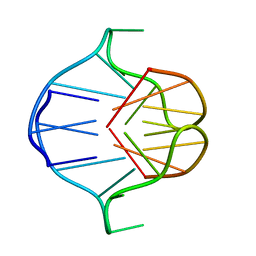 | |
1A8K
 
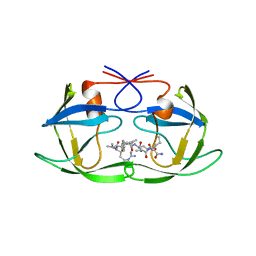 | | CRYSTALLOGRAPHIC ANALYSIS OF HUMAN IMMUNODEFICIENCY VIRUS 1 PROTEASE WITH AN ANALOG OF THE CONSERVED CA-P2 SUBSTRATE: INTERACTIONS WITH FREQUENTLY OCCURRING GLUTAMIC ACID RESIDUE AT P2' POSITION OF SUBSTRATES | | Descriptor: | HIV PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Weber, I.T, Wu, J, Adomat, J, Harrison, R.W, Kimmel, A.R, Wondrak, E.M, Louis, J.M. | | Deposit date: | 1998-03-27 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of human immunodeficiency virus 1 protease with an analog of the conserved CA-p2 substrate -- interactions with frequently occurring glutamic acid residue at P2' position of substrates.
Eur.J.Biochem., 249, 1997
|
|
1B5T
 
 | | ESCHERICHIA COLI METHYLENETETRAHYDROFOLATE REDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MERCURY (II) ION, PROTEIN (METHYLENETETRAHYDROFOLATE REDUCTASE) | | Authors: | Guenther, B.D, Sheppard, C.A, Tran, P, Rozen, R, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure and properties of methylenetetrahydrofolate reductase from Escherichia coli suggest how folate ameliorates human hyperhomocysteinemia.
Nat.Struct.Biol., 6, 1999
|
|
1BIO
 
 | | HUMAN COMPLEMENT FACTOR D IN COMPLEX WITH ISATOIC ANHYDRIDE INHIBITOR | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL, ISATOIC ANHYDRIDE | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
1BIT
 
 | |
1BIL
 
 | | CRYSTALLOGRAPHIC STUDIES ON THE BINDING MODES OF P2-P3 BUTANEDIAMIDE RENIN INHIBITORS | | Descriptor: | (2S)-2-[(2-amino-1,3-thiazol-4-yl)methyl]-N~1~-[(1S,2R,3R)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methylhexyl]-N~4~-[2-(d imethylamino)-2-oxoethyl]-N~4~-[(1S)-1-phenylethyl]butanediamide, Renin | | Authors: | Tong, L. | | Deposit date: | 1995-09-27 | | Release date: | 1996-01-29 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies on the binding modes of P2-P3 butanediamide renin inhibitors.
J.Biol.Chem., 270, 1995
|
|
1BG0
 
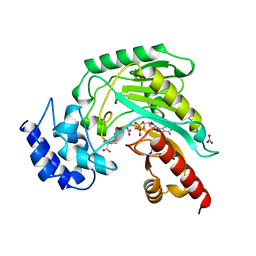 | | TRANSITION STATE STRUCTURE OF ARGININE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE KINASE, D-ARGININE, ... | | Authors: | Zhou, G, Somasundaram, T, Blanc, E, Parthasarathy, G, Ellington, W.R, Chapman, M.S. | | Deposit date: | 1998-06-03 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Transition state structure of arginine kinase: implications for catalysis of bimolecular reactions.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BHB
 
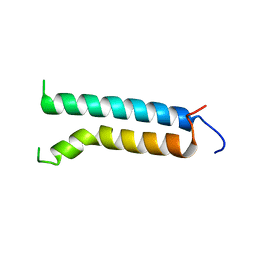 | | Three-dimensional structure of (1-71) bacterioopsin solubilized in methanol-chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy | | Descriptor: | BACTERIORHODOPSIN | | Authors: | Orekhov, V.Y, Pervushin, K.V, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | Deposit date: | 1993-10-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
