3DK1
 
 | | Wild Type HIV-1 Protease with potent Antiviral inhibitor GRL-0105A | | Descriptor: | (4aR,6r,7aS)-hexahydro-4aH-cyclopenta[b][1,4]dioxin-6-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.F, Weber, I.T. | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Potent HIV-1 protease inhibitors incorporating meso-bicyclic urethanes as P2-ligands: structure-based design, synthesis, biological evaluation and protein-ligand X-ray studies
Org.Biomol.Chem., 6, 2008
|
|
3DO7
 
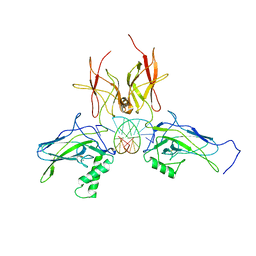 | | X-ray structure of a NF-kB p52/RelB/DNA complex | | Descriptor: | 5'-D(*DCP*DGP*DGP*DGP*DAP*DAP*DTP*DTP*DCP*DCP*DC)-3', Avian reticuloendotheliosis viral (V-rel) oncogene related B, Nuclear factor NF-kappa-B p100 subunit | | Authors: | Fusco, A, Huang, D.B, Miller, D, Vu, D, Ghosh, G. | | Deposit date: | 2008-07-03 | | Release date: | 2009-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | NF-kapppaB p52:RelB heterodimer uses different binding modes to recognize different kappaB DNA
TO BE PUBLISHED
|
|
4LSJ
 
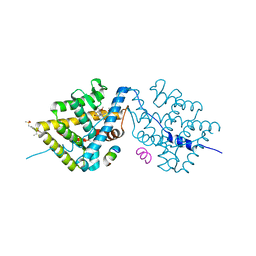 | | Crystal Structure of the Glucocorticoid Receptor Ligand Binding Domain Bound to a Dibenzoxapine Sulfonamide | | Descriptor: | D30 peptide, Glucocorticoid receptor, N-{3-[(1Z)-1-(10-methoxydibenzo[b,e]oxepin-11(6H)-ylidene)propyl]phenyl}methanesulfonamide | | Authors: | Carson, M, Luz, J.G, Clawson, D, Coghlan, M. | | Deposit date: | 2013-07-22 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Glucocorticoid receptor modulators informed by crystallography lead to a new rationale for receptor selectivity, function, and implications for structure-based design.
J.Med.Chem., 57, 2014
|
|
3MHR
 
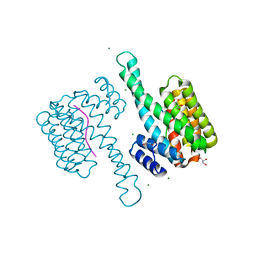 | | 14-3-3 sigma in complex with YAP pS127-peptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schumacher, B, Skwarczynska, M, Rose, R, Ottmann, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of a 14-3-3[sigma]-YAP phosphopeptide complex at 1.15 A resolution
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1NUN
 
 | | Crystal Structure Analysis of the FGF10-FGFR2b Complex | | Descriptor: | Fibroblast growth factor-10, POLYETHYLENE GLYCOL (N=34), SULFATE ION, ... | | Authors: | Yeh, B.K, Igarashi, M, Eliseenkova, A.V, Plotnikov, A.N, Sher, I, Ron, D, Aaronson, S.A, Mohammadi, M. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis by which alternative splicing confers
specificity in fibroblast growth factor receptors.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1D58
 
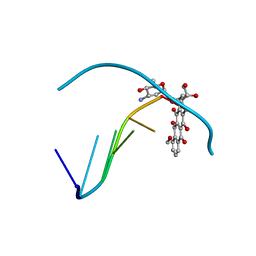 | | THE MOLECULAR STRUCTURE OF A 4'-EPIADRIAMYCIN COMPLEX WITH D(TGATCA) AT 1.7 ANGSTROM RESOLUTION-COMPARISON WITH THE STRUCTURE OF 4'-EPIADRIAMYCIN D(TGTACA) AND D(CGATCG) COMPLEXES | | Descriptor: | 4'-EPIDOXORUBICIN, DNA (5'-D(*TP*GP*AP*TP*CP*A)-3') | | Authors: | Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1992-02-20 | | Release date: | 1992-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular structure of a 4'-epiadriamycin complex with d(TGATCA) at 1.7A resolution: comparison with the structure of 4'-epiadriamycin d(TGTACA) and d(CGATCG) complexes.
Nucleic Acids Res., 20, 1992
|
|
2WMF
 
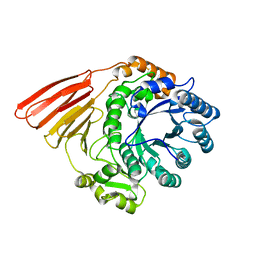 | | Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in its native form. | | Descriptor: | FUCOLECTIN-RELATED PROTEIN | | Authors: | Higgins, M.A, Whitworth, G.E, El Warry, N, Randriantsoa, M, Samain, E, Burke, R.D, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Recognition and Hydrolysis of Host Carbohydrate-Antigens by Streptococcus Pneumoniae Family 98 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
2H04
 
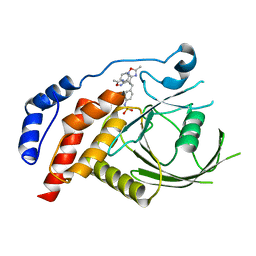 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Protein tyrosine phosphatase, receptor type, B,, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M, Gray, J.L, Peters, K.G, Maier, M.B, Amarasinghe, K.D, Clark, C.M, Nichols, R. | | Deposit date: | 2006-05-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of potent, non-peptidic inhibitors of HPTPbeta.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HD3
 
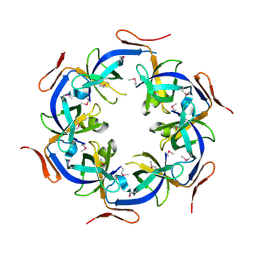 | | Crystal Structure of the Ethanolamine Utilization Protein EutN from Escherichia coli, NESG Target ER316 | | Descriptor: | Ethanolamine utilization protein eutN | | Authors: | Forouhar, F, Zhang, W, Jayaraman, S, Zhao, L, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-19 | | Release date: | 2006-08-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
4ACV
 
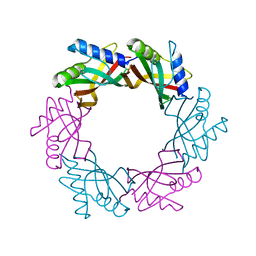 | |
1CGE
 
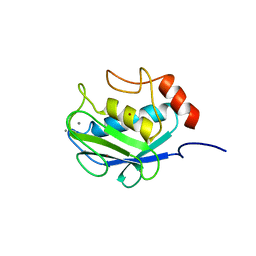 | | CRYSTAL STRUCTURES OF RECOMBINANT 19-KDA HUMAN FIBROBLAST COLLAGENASE COMPLEXED TO ITSELF | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, ZINC ION | | Authors: | Lovejoy, B, Hassell, A.M, Luther, M.A, Weigl, D, Jordan, S.R. | | Deposit date: | 1994-02-03 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of recombinant 19-kDa human fibroblast collagenase complexed to itself.
Biochemistry, 33, 1994
|
|
4EKA
 
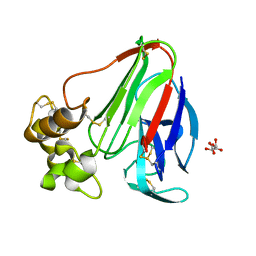 | | Final Thaumatin Structure for Radiation Damage Experiment at 25 K | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | Deposit date: | 2012-04-09 | | Release date: | 2012-08-29 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4KZG
 
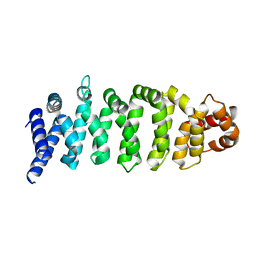 | |
1D8G
 
 | | ULTRAHIGH RESOLUTION CRYSTAL STRUCTURE OF B-DNA DECAMER D(CCAGTACTGG) | | Descriptor: | 5'-D(*CP*CP*AP*GP*TP*AP*CP*TP*GP*GP*)-3', CALCIUM ION | | Authors: | Kielkopf, C.L, Ding, S, Kuhn, P, Rees, D.C. | | Deposit date: | 1999-10-23 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.74 Å) | | Cite: | Conformational flexibility of B-DNA at 0.74 A resolution: d(CCAGTACTGG)(2).
J.Mol.Biol., 296, 2000
|
|
1UN3
 
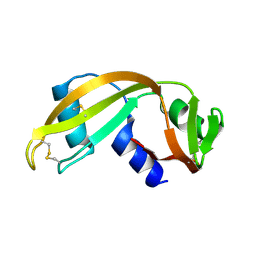 | |
3RN3
 
 | | SEGMENTED ANISOTROPIC REFINEMENT OF BOVINE RIBONUCLEASE A BY THE APPLICATION OF THE RIGID-BODY TLS MODEL | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Howlin, B, Moss, D.S, Harris, G.W, Palmer, R.A. | | Deposit date: | 1991-10-30 | | Release date: | 1991-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Segmented anisotropic refinement of bovine ribonuclease A by the application of the rigid-body TLS model.
Acta Crystallogr.,Sect.A, 45, 1989
|
|
3BJ9
 
 | | Crystal structure of the Surrogate Light Chain Variable Domain VpreBJ | | Descriptor: | 1,2-ETHANEDIOL, Immunoglobulin iota chain, Immunoglobulin lambda-like polypeptide 1 | | Authors: | Morstadt, L.M, Bohm, A.A, Stollar, B.D, Baleja, J.D. | | Deposit date: | 2007-12-03 | | Release date: | 2008-03-04 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering and characterization of a single chain surrogate light chain variable domain.
Protein Sci., 17, 2008
|
|
3UN5
 
 | | Bacillus cereus phosphopentomutase T85E variant | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3R66
 
 | | Crystal structure of human ISG15 in complex with NS1 N-terminal region from influenza virus B, Northeast Structural Genomics Consortium Target IDs HX6481, HR2873, and OR2 | | Descriptor: | Non-structural protein 1, Ubiquitin-like protein ISG15 | | Authors: | Guan, R, Ma, L.C, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-21 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the complex of ISG15 and Influenza B virus NS1 Protein: implications for the mechanism of SN1-mediated inhibition of ISG15 conjugation
To be Published
|
|
3R8B
 
 | |
1LAJ
 
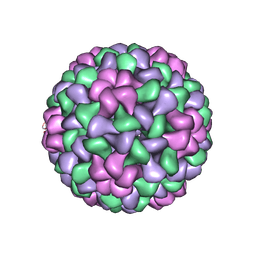 | | The Structure of Tomato Aspermy Virus by X-Ray Crystallography | | Descriptor: | 5'-R(*AP*AP*A)-3', MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lucas, R.W, Larson, S.B, Canady, M.A, McPherson, A. | | Deposit date: | 2002-03-28 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structure of Tomato Aspermy Virus by X-Ray Crystallography
J.STRUCT.BIOL., 139, 2002
|
|
4DEG
 
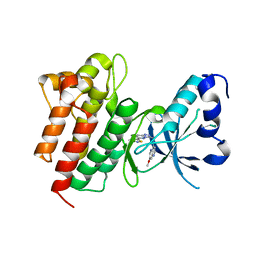 | | Crystal structure of c-Met in complex with triazolopyridazine inhibitor 2 | | Descriptor: | 7-methoxy-N-{[6-(3-methyl-1,2-thiazol-5-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]methyl}-1,5-naphthyridin-4-amine, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Bellon, S.F, Long, A.M. | | Deposit date: | 2012-01-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of a potent and selective triazolopyridinone series of c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4PPX
 
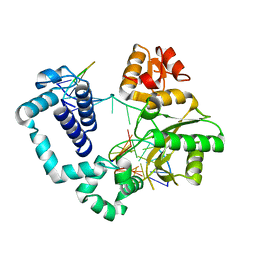 | | DNA Polymerase Beta E295K with Spiroiminodihydantoin in Templating Position | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*(SDH)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*G)-3', ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2014-02-27 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of DNA Polymerase beta with DNA Containing the Base Lesion Spiroiminodihydantoin in a Templating Position.
Biochemistry, 53, 2014
|
|
2BLG
 
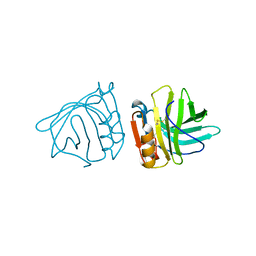 | | STRUCTURAL BASIS OF THE TANFORD TRANSITION OF BOVINE BETA-LACTOGLOBULIN FROM CRYSTAL STRUCTURES AT THREE PH VALUES; PH 8.2 | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Qin, B.Y, Bewley, M.C, Creamer, L.K, Baker, H.M, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1999-01-27 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis of the Tanford transition of bovine beta-lactoglobulin.
Biochemistry, 37, 1998
|
|
3UNY
 
 | | Bacillus cereus phosphopentomutase T85E variant soaked with glucose 1,6-bisphosphate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
