7SCV
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL17 | | Descriptor: | C-C motif chemokine 17, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
7SCT
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL16 | | Descriptor: | C-C motif chemokine 16, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
7SCS
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL11 | | Descriptor: | Eotaxin, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
7SCU
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of Tick Evasin EVA-AAM1001 complexed to human chemokine CCL7
To Be Published
|
|
7PUA
 
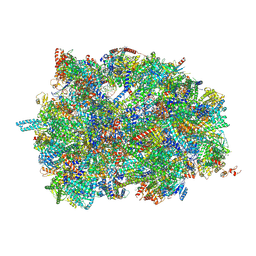 | | Middle assembly intermediate of the Trypanosoma brucei mitoribosomal small subunit | | Descriptor: | 30S Ribosomal protein S17, putative, 30S ribosomal protein S8, ... | | Authors: | Lenarcic, T, Leibundgut, M, Saurer, M, Ramrath, D.J.F, Fluegel, T, Boehringer, D, Ban, N. | | Deposit date: | 2021-09-29 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mitoribosomal small subunit maturation involves formation of initiation-like complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PUB
 
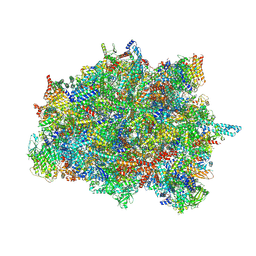 | | Late assembly intermediate of the Trypanosoma brucei mitoribosomal small subunit | | Descriptor: | 30S Ribosomal protein S17, putative, 30S ribosomal protein S8, ... | | Authors: | Lenarcic, T, Leibundgut, M, Saurer, M, Ramrath, D.J.F, Fluegel, T, Boehringer, D, Ban, N. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mitoribosomal small subunit maturation involves formation of initiation-like complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SCG
 
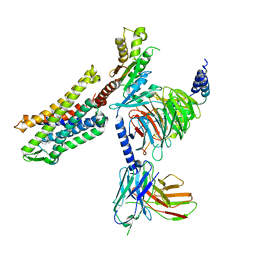 | | FH210 bound Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | (2E)-N-[(2S)-2-(dimethylamino)-3-(4-hydroxyphenyl)propyl]-3-(naphthalen-1-yl)prop-2-enamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, H, Kobilka, B. | | Deposit date: | 2021-09-28 | | Release date: | 2022-04-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-Based Evolution of G Protein-Biased mu-Opioid Receptor Agonists.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7PU5
 
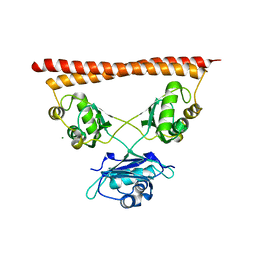 | | Structure of SFPQ-NONO complex | | Descriptor: | MAGNESIUM ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Fribourg, S. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Crystal structure of SFPQ-NONO heterodimer.
Biochimie, 198, 2022
|
|
7PTY
 
 | | Delta-latroinsectotoxin dimer | | Descriptor: | Delta-latroinsectotoxin-Lt1a | | Authors: | Chen, M, Gatsogiannis, C. | | Deposit date: | 2021-09-27 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.63 Å) | | Cite: | Molecular architecture of black widow spider neurotoxins.
Nat Commun, 12, 2021
|
|
7SCE
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-2 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7SCD
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-1 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7PT7
 
 | | Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, state I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 7, ... | | Authors: | Saleh, A, Noguchi, Y, Aramayo, R, Ivanova, M.E, Speck, C. | | Deposit date: | 2021-09-26 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structural basis of Cdc7-Dbf4 kinase dependent targeting and phosphorylation of the MCM2-7 double hexamer.
Nat Commun, 13, 2022
|
|
7PT6
 
 | | Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 7, DDK kinase regulatory subunit DBF4, ... | | Authors: | Saleh, A, Noguchi, Y, Aramayo, R, Ivanova, M.E, Speck, C. | | Deposit date: | 2021-09-26 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structural basis of Cdc7-Dbf4 kinase dependent targeting and phosphorylation of the MCM2-7 double hexamer.
Nat Commun, 13, 2022
|
|
7VIG
 
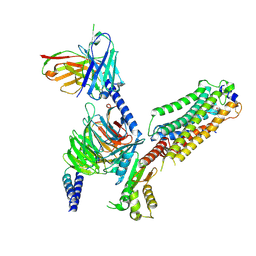 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307 | | Descriptor: | 1-[[2-fluoranyl-4-[5-[4-(2-methylpropyl)phenyl]-1,2,4-oxadiazol-3-yl]phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VIE
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VIF
 
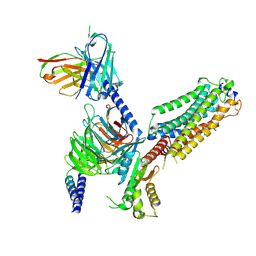 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with (S)-FTY720-P | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VIH
 
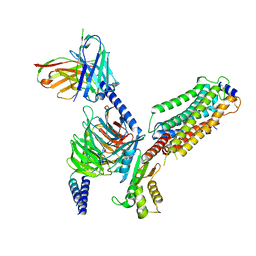 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307 | | Descriptor: | 1-[[2-fluoranyl-4-[5-[4-(2-methylpropyl)phenyl]-1,2,4-oxadiazol-3-yl]phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SBF
 
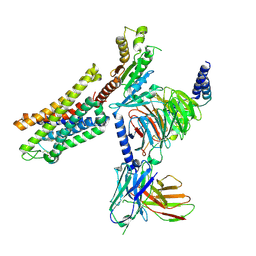 | | PZM21 bound Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Huang, W, Qu, Q, Wang, H, Skiniotis, G, Kobilka, B. | | Deposit date: | 2021-09-24 | | Release date: | 2022-04-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-Based Evolution of G Protein-Biased mu-Opioid Receptor Agonists.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7PSX
 
 | | Structure of HOXB13 bound to hydroxymethylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*5HCP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2021-09-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of HOXB13 bound to hydroxymethylated DNA
To Be Published
|
|
7SBH
 
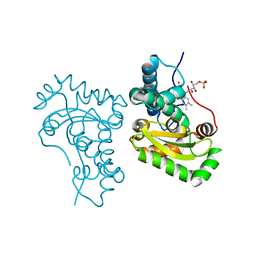 | | Crystal structure of the iron superoxide dismutase from Acinetobacter sp. Ver3 | | Descriptor: | FE (III) ION, FLAVIN MONONUCLEOTIDE, Superoxide dismutase | | Authors: | Steimbruch, B.A, Albanesi, D, Repizo, G.D, Lisa, M.N. | | Deposit date: | 2021-09-24 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The distinctive roles played by the superoxide dismutases of the extremophile Acinetobacter sp. Ver3.
Sci Rep, 12, 2022
|
|
7PSL
 
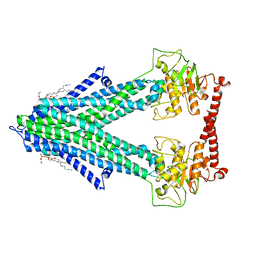 | | S. cerevisiae Atm1 in MSP1D1 nanodiscs in nucleotide-free state | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, Iron-sulfur clusters transporter ATM1, mitochondrial, ... | | Authors: | Ellinghaus, T.L, Kuehlbrandt, W. | | Deposit date: | 2021-09-23 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes in the yeast mitochondrial ABC transporter Atm1 during the transport cycle.
Sci Adv, 7, 2021
|
|
7PSM
 
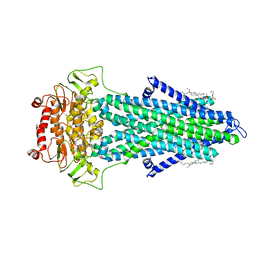 | | S. cerevisiae Atm1 in MSP1D1 nanodiscs with bound AMP-PNP and Mg2+ | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, Iron-sulfur clusters transporter ATM1, mitochondrial, ... | | Authors: | Ellinghaus, T.L, Kuehlbrandt, W. | | Deposit date: | 2021-09-23 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conformational changes in the yeast mitochondrial ABC transporter Atm1 during the transport cycle.
Sci Adv, 7, 2021
|
|
7PSN
 
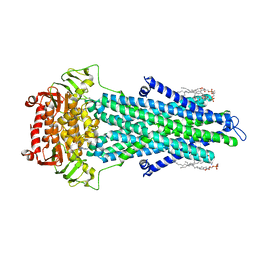 | |
7PSU
 
 | | Structure of protein kinase CK2alpha mutant K198R associated with the Okur-Chung Neurodevelopmental Syndrome | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Werner, C, Gast, A, Lindenblatt, D, Nickelsen, K, Niefind, K, Jose, J, Hochscherf, J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and Enzymological Evidence for an Altered Substrate Specificity in Okur-Chung Neurodevelopmental Syndrome Mutant CK2 alpha Lys198Arg.
Front Mol Biosci, 9, 2022
|
|
7SA7
 
 | |
