3WUE
 
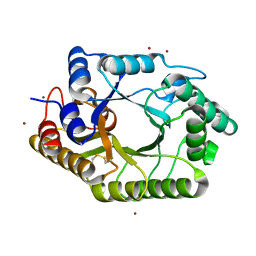 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3KS0
 
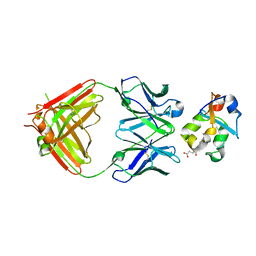 | | Crystal structure of the heme domain of flavocytochrome b2 in complex with Fab B2B4 | | Descriptor: | Cytochrome b2, mitochondrial, Fragment Antigen Binding B2B4, ... | | Authors: | Golinelli-Pimpaneau, B, Lederer, F, Le, K.H.D. | | Deposit date: | 2009-11-20 | | Release date: | 2010-05-26 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for the functional importance of the heme domain mobility in flavocytochrome b2.
J.Mol.Biol., 400, 2010
|
|
4QCI
 
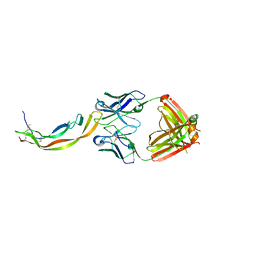 | | PDGF-B blocking antibody bound to PDGF-BB | | Descriptor: | Platelet-derived growth factor subunit B, anti-PDGF-BB antibody - Light Chain, anti-PDGF-BB antibody - Heavy chain | | Authors: | Kuai, J, Mosyak, L, Tam, M, LaVallie, E, Pullen, N, Carven, G. | | Deposit date: | 2014-05-12 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Binding Mode of Action of a Blocking Anti-Platelet-Derived Growth Factor (PDGF)-B Monoclonal Antibody, MOR8457, Reveals Conformational Flexibility and Avidity Needed for PDGF-BB To Bind PDGF Receptor-beta.
Biochemistry, 54, 2015
|
|
3EXX
 
 | |
2Y7A
 
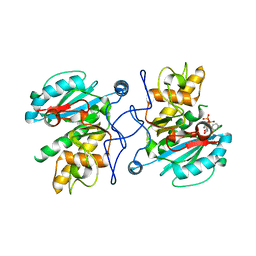 | | Crystal structure of unliganded GTB P156L | | Descriptor: | ABO GLYCOSYLTRANSFERASE, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Weadge, J, Palcic, M.M, Henriksen, A. | | Deposit date: | 2011-01-31 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and Biochemical Characterization of the Human Blood Group a and B Galactosyltransferases Possessing the Pro156Leu Mutation
To be Published
|
|
3HC7
 
 | |
1XFB
 
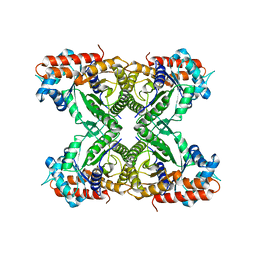 | | Human Brain Fructose 1,6-(bis)phosphate Aldolase (C isozyme) | | Descriptor: | Aldolase C | | Authors: | Arakaki, T.L, Pezza, J.A, Cronin, M.A, Hopkins, C.E, Zimmer, D.B, Tolan, D.R, Allen, K.N. | | Deposit date: | 2004-09-14 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human brain fructose 1,6-(bis)phosphate aldolase: linking isozyme structure with function
Protein Sci., 13, 2004
|
|
4DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
1FX9
 
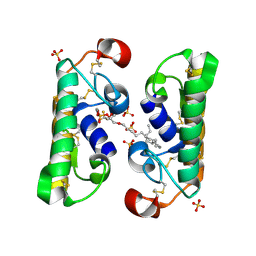 | | CARBOXYLIC ESTER HYDROLASE COMPLEX (DIMERIC PLA2 + MJ33 INHIBITOR + SULPHATE IONS) | | Descriptor: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PHOSPHOLIPASE A2, ... | | Authors: | Pan, Y.H, Epstein, T.M, Jain, M.K, Bahnson, B.J. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Five coplanar anion binding sites on one face of phospholipase A2: relationship to interface binding.
Biochemistry, 40, 2001
|
|
5XF3
 
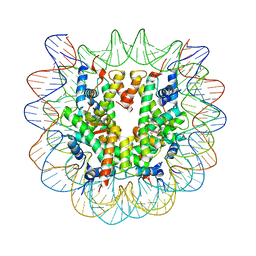 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (R,R-configuration) | | Descriptor: | (1R,2R)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
4AE6
 
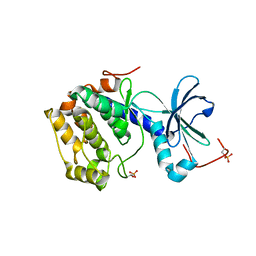 | | Structure and Function of the Human Sperm-Specific Isoform of Protein Kinase A (PKA) Catalytic Subunit Calpha 2 | | Descriptor: | ACETATE ION, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA 2 | | Authors: | Hereng, T.H, Backe, P.H, Kahmann, J, Scheich, C, Bjoras, M, Skalhegg, B.S, Rosendal, K.R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of the Human Sperm-Specific Isoform of Protein Kinase a (Pka) Catalytic Subunit Calpha2
J.Struct.Biol., 178, 2012
|
|
3GPM
 
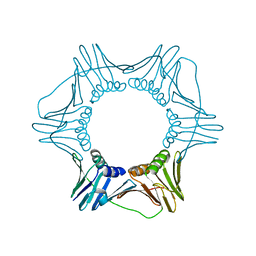 | | Structure of the trimeric form of the E113G PCNA mutant protein | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Freudenthal, B.D, Gakhar, L, Ramaswamy, S, Washington, M.T. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A charged residue at the subunit interface of PCNA promotes trimer formation by destabilizing alternate subunit interactions.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4BQ7
 
 | | Crystal structure of the RGMB-Neo1 complex form 2 | | Descriptor: | NEOGENIN, RGM DOMAIN FAMILY MEMBER B | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (6.601 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
3HAM
 
 | | Structure of the gentamicin-APH(2")-IIa complex | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, Aminoglycoside phosphotransferase, GLYCEROL | | Authors: | Young, P.G, Baker, E.N, Vakulenko, S.B, Smith, C.A. | | Deposit date: | 2009-05-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2''-phosphotransferase-IIa [APH(2'')-IIa] provide insights into substrate selectivity in the APH(2'') subfamily.
J.Bacteriol., 191, 2009
|
|
3HE8
 
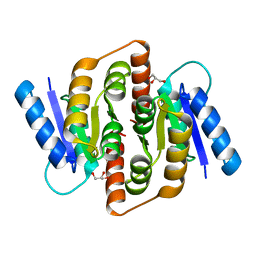 | | Structural study of Clostridium thermocellum Ribose-5-Phosphate Isomerase B | | Descriptor: | GLYCEROL, Ribose-5-phosphate isomerase | | Authors: | Kang, L.W, Kim, J.K, Jung, J.H, Hong, M.K. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
4U20
 
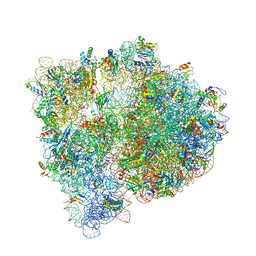 | | Crystal structure of the E. coli ribosome bound to flopristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
1UY2
 
 | |
3HEE
 
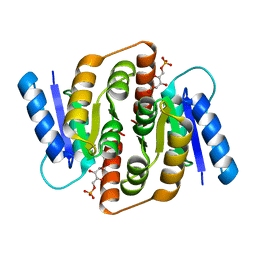 | | Structural study of Clostridium thermocellum Ribose-5-Phosphate Isomerase B and ribose-5-phosphate | | Descriptor: | RIBOSE-5-PHOSPHATE, Ribose-5-phosphate isomerase | | Authors: | Kang, L.W, Kim, J.K, Jung, J.H, Hong, M.K. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
1PVE
 
 | |
1ZJ1
 
 | | Crystal Structure of Human Galactosyltransferase (GTB) Complexed with N-acetyllactosamine | | Descriptor: | ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase), ... | | Authors: | Letts, J.A, Rose, N.L, Fang, Y.R, Barry, C.H, Borisova, S.N, Seto, N.O, Palcic, M.M, Evans, S.V. | | Deposit date: | 2005-04-27 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differential Recognition of the Type I and II H Antigen Acceptors by the Human ABO(H) Blood Group A and B Glycosyltransferases.
J.Biol.Chem., 281, 2006
|
|
4FK6
 
 | | JAK1 kinase (JH1 domain) in complex with compound 72 | | Descriptor: | N-({1-[(1R,2R,4S)-bicyclo[2.2.1]hept-2-yl]-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridin-2-yl}methyl)methanesulfonamide, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2012-06-12 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based discovery of C-2 substituted imidazo-pyrrolopyridine JAK1 inhibitors with improved selectivity over JAK2.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1QK9
 
 | | The solution structure of the domain from MeCP2 that binds to methylated DNA | | Descriptor: | METHYL-CPG-BINDING PROTEIN 2 | | Authors: | Wakefield, R.I.D, Smith, B.O, Nan, X, Free, A, Soteriou, A, Uhrin, D, Bird, A.P, Barlow, P.N. | | Deposit date: | 1999-07-12 | | Release date: | 1999-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Domain from Mecp2 that Binds to Methylated DNA
J.Mol.Biol., 291, 1999
|
|
3CWV
 
 | | Crystal structure of B-subunit of the DNA gyrase from Myxococcus xanthus | | Descriptor: | DNA gyrase, B subunit, truncated | | Authors: | Ramagopal, U.A, Toro, R, Meyer, A.J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-22 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of B-subunit of the DNA gyrase from Myxococcus xanthus.
To be published
|
|
2M0P
 
 | | Solution structure of the tenth complement type repeat of human megalin | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 2 | | Authors: | Dagil, R, Kragelund, B. | | Deposit date: | 2012-11-01 | | Release date: | 2013-01-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Gentamicin binds to the megalin receptor as a competitive inhibitor using the common ligand binding motif of complement type repeats: insight from the nmr structure of the 10th complement type repeat domain alone and in complex with gentamicin.
J.Biol.Chem., 288, 2013
|
|
1RT3
 
 | | AZT DRUG RESISTANT HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH 1051U91 | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-06-29 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3'-Azido-3'-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
