1CQG
 
 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN REF-1 (RESIDUES 59-71 OF THE P50 SUBUNIT OF NFKB), NMR, 31 STRUCTURES | | Descriptor: | REF-1 PEPTIDE, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1996-04-02 | | Release date: | 1996-08-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal.
Structure, 4, 1996
|
|
9HCJ
 
 | | D. discoideum nuclear pore complex | | Descriptor: | ELYS-like domain-containing protein, NDC1, Nuclear pore complex protein, ... | | Authors: | Obarska-Kosinska, A, Hoffmann, P.C, Beck, M. | | Deposit date: | 2024-11-10 | | Release date: | 2024-12-25 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Nuclear pore permeability and fluid flow are modulated by its dilation state.
Mol.Cell, 85, 2025
|
|
9D8P
 
 | | Focused map of Cryo-EM structure of Ubiquitin C-degron bound to KLHDC10-EloB/C | | Descriptor: | Elongin-B, Elongin-C, Kelch domain-containing protein 10, ... | | Authors: | Chittori, S, Scott, D.C, Schulman, B.A. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-20 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for C-degron selectivity across KLHDCX family E3 ubiquitin ligases.
Nat Commun, 15, 2024
|
|
8J07
 
 | |
3MHH
 
 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
5WG6
 
 | | Human Polycomb Repressive Complex 2 in complex with GSK126 inhibitor | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine N-methyltransferase EZH2,Polycomb protein SUZ12 (E.C.2.1.1.43) chimera, Polycomb protein EED, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
3MHS
 
 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to ubiquitin aldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
7K5Y
 
 | |
6UXW
 
 | | SWI/SNF nucleosome complex with ADP-BeFx | | Descriptor: | 601 sequence bottom strand, 601 sequence top strand, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | He, Y, Han, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (8.96 Å) | | Cite: | Cryo-EM structure of SWI/SNF complex bound to a nucleosome.
Nature, 579, 2020
|
|
3KJL
 
 | | Sgf11:Sus1 complex | | Descriptor: | Protein SUS1, SAGA-associated factor 11 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2009-11-03 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1.
J.Biol.Chem., 285, 2010
|
|
6V0O
 
 | | PRMT5 bound to the PBM peptide from pICln | | Descriptor: | ACETYL GROUP, Methylosome protein 50, PBM peptide, ... | | Authors: | McMillan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
8B3I
 
 | | CRL4CSA-E2-Ub (state 2) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | C(N)RL4CSA-E2-Ub (state 2)
To Be Published
|
|
6UXX
 
 | | PRMT5:MEP50 Complexed with Allosteric Inhibitor Compound 1a | | Descriptor: | (5R)-2-amino-5-(4-methoxyphenyl)-3-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-yl]-3,5-dihydro-4H-imidazol-4-one, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Palte, R.L, Schneider, S.E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Acs Med.Chem.Lett., 11, 2020
|
|
6V0P
 
 | | PRMT5 complex bound to covalent PBM inhibitor BRD6711 | | Descriptor: | 2-(5-chloro-6-oxopyridazin-1(6H)-yl)-N-(4-methyl-3-sulfamoylphenyl)acetamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | McMillan, B.J, McKinney, D.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the PRMT5-Substrate Adaptor Interaction.
J.Med.Chem., 64, 2021
|
|
4XMN
 
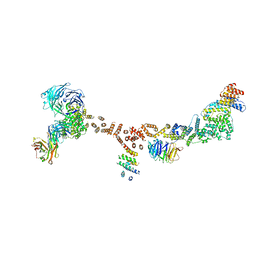 | | Structure of the yeast coat nucleoporin complex, space group P212121 | | Descriptor: | Antibody 87 heavy chain, Antibody 87 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
3KIK
 
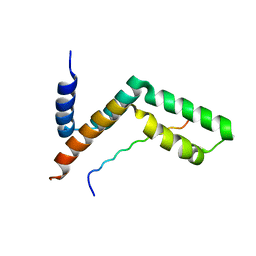 | | Sgf11:Sus1 complex | | Descriptor: | Protein SUS1, SAGA-associated factor 11 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2009-11-02 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1
J.Biol.Chem., 285, 2010
|
|
4XMM
 
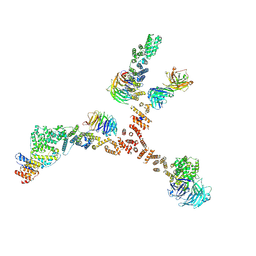 | | Structure of the yeast coat nucleoporin complex, space group C2 | | Descriptor: | Antibody 57 heavy chain, Antibody 57 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (7.384 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4F52
 
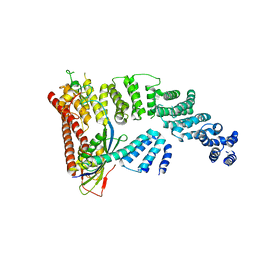 | | Structure of a Glomulin-RBX1-CUL1 complex | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, Glomulin, ... | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2012-05-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a Glomulin-RBX1-CUL1 Complex: Inhibition of a RING E3 Ligase through Masking of Its E2-Binding Surface.
Mol.Cell, 47, 2012
|
|
5A5B
 
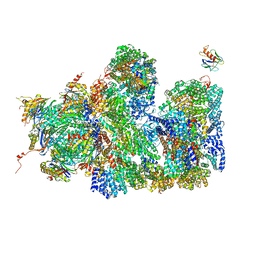 | | Structure of the 26S proteasome-Ubp6 complex | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Aufderheide, A, Beck, F, Stengel, F, Hartwig, M, Schweitzer, A, Pfeifer, G, Goldberg, A.L, Sakata, E, Baumeister, W, Foerster, F. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-22 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structural Characterization of the Interaction of Ubp6 with the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8YYV
 
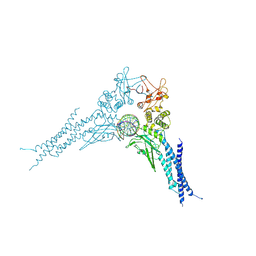 | | A dimeric STAT1-DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*TP*G)-3'), Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Sugiyama, A, Minami, M, Sugita, Y, Ose, T. | | Deposit date: | 2024-04-04 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural analysis reveals how tetrameric tyrosine-phosphorylated STAT1 is targeted by the rabies virus P-protein.
Sci.Signal., 18, 2025
|
|
8YYU
 
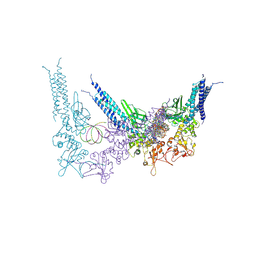 | | A tetrameric STAT1-DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*TP*G)-3'), Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Sugiyama, A, Minami, M, Sugita, Y, Ose, T. | | Deposit date: | 2024-04-04 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural analysis reveals how tetrameric tyrosine-phosphorylated STAT1 is targeted by the rabies virus P-protein.
Sci.Signal., 18, 2025
|
|
9DDE
 
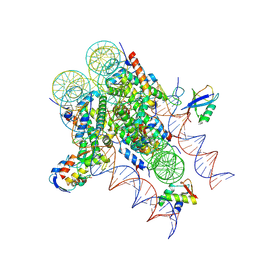 | | ncPRC1RYBP bound to H2AK119Ub/H1.4 chromatosome | | Descriptor: | DNA (187-MER), E3 ubiquitin-protein ligase RING2, Histone H1.4, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-08-28 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
8VAT
 
 | |
8F86
 
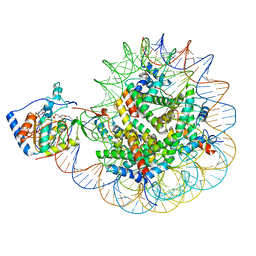 | | SIRT6 bound to an H3K9Ac nucleosome | | Descriptor: | DNA (148-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Markert, J, Whedon, S, Wang, Z, Cole, P, Farnung, L. | | Deposit date: | 2022-11-21 | | Release date: | 2023-04-05 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Sirtuin 6-Catalyzed Nucleosome Deacetylation.
J.Am.Chem.Soc., 145, 2023
|
|
8X77
 
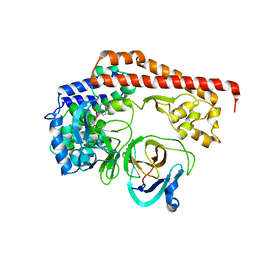 | | Enterovirus proteinase with host factor | | Descriptor: | 2A protein, Actin-histidine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2023-11-23 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Nat Commun, 15, 2024
|
|
