7QQL
 
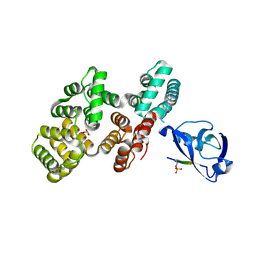 | | The PDZ domain of SNTG2 complexed with the phosphorylated PDZ-binding motif of RSK1 | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-2-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WU7
 
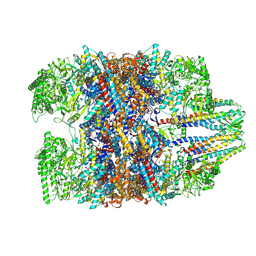 | | Prefoldin-tubulin-TRiC complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Prefoldin subunit 1, Prefoldin subunit 2, ... | | Authors: | Gestaut, D, Zhao, Y, Park, J, Ma, B, Leitner, A, Collier, M, Pintilie, G, Roh, S.-H, Chiu, W, Frydman, J. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
6WKR
 
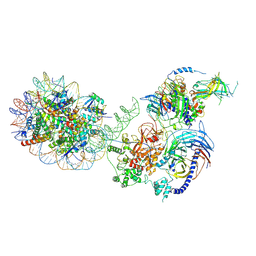 | | PRC2-AEBP2-JARID2 bound to H2AK119ub1 nucleosome | | Descriptor: | DNA (314-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Kasinath, V, Nogales, E, Beck, C, Sauer, P, Poepsel, S, Kosmatka, J, Faini, M, Toso, D, Aebersold, R. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-03 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications.
Science, 371, 2021
|
|
4LJX
 
 | |
4W9D
 
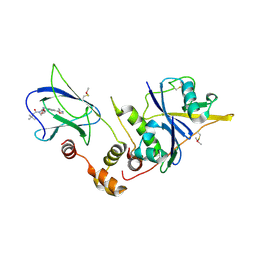 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methyloxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
3KAB
 
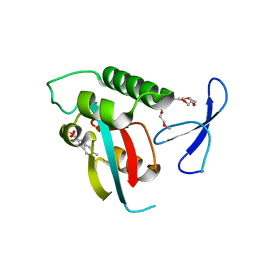 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 6-methyl-1H-indole-2-carboxylic acid, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4L4M
 
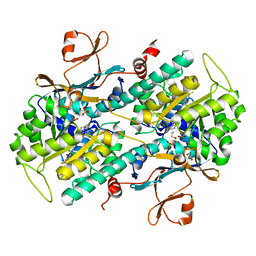 | | Structural Analysis of a Phosphoribosylated Inhibitor in Complex with Human Nicotinamide Phosphoribosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-7-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Ho, Y, Zak, M, Liu, Y, Yuen, P, Zheng, X, Dragovich, S.P, Wang, W. | | Deposit date: | 2013-06-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
4W9C
 
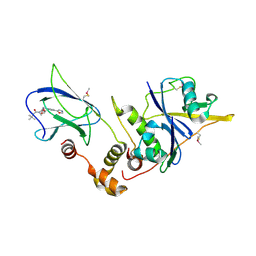 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(oxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 2) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4W9H
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-acetamido-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 7) | | Descriptor: | N-acetyl-3-methyl-L-valyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4W9E
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(thiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 4) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4LVG
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | (1S,2S)-N-[4-(phenylsulfonyl)phenyl]-2-(pyridin-3-yl)cyclopropanecarboxamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4W9J
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-((S)-2-acetamido-4-methylpentanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 13) | | Descriptor: | N-acetyl-L-leucyl-3-methyl-L-valyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4W9L
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-((S)-2-acetamido-3,3-dimethylbutanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 15) | | Descriptor: | N-acetyl-3-methyl-L-valyl-3-methyl-L-valyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
7SOY
 
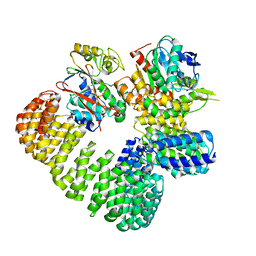 | | The structure of the PP2A-B56gamma1 holoenzyme-PME-1 complex | | Descriptor: | Isoform Gamma-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, Protein phosphatase methylesterase 1, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Li, Y, Balakrishnan, V.K, Rowse, M, Novikova, I.V, Xing, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Coupling to short linear motifs creates versatile PME-1 activities in PP2A holoenzyme demethylation and inhibition.
Elife, 11, 2022
|
|
6OBN
 
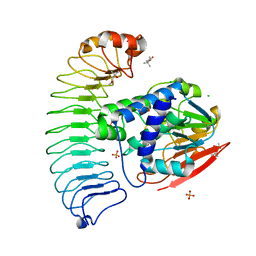 | | The crystal structure of coexpressed SDS22:PP1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5MPC
 
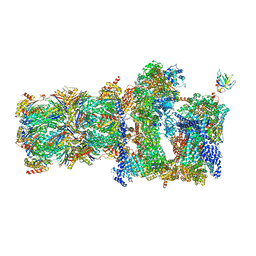 | | 26S proteasome in presence of BeFx (s4) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8KCY
 
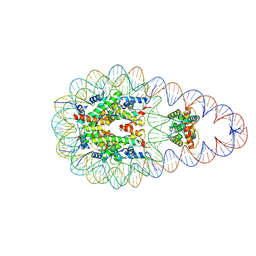 | | Structure of nucleosome complexed with two DEK molecules | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kujirai, T, Echigoya, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-08-08 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into how DEK nucleosome binding facilitates H3K27 trimethylation in chromatin.
Nat.Struct.Mol.Biol., 2025
|
|
8KD1
 
 | | Structure of nucleosome complexed with one DEK molecule | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kujirai, T, Echigoya, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-08-08 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into how DEK nucleosome binding facilitates H3K27 trimethylation in chromatin.
Nat.Struct.Mol.Biol., 2025
|
|
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
9JNV
 
 | | Structure of isw1-nucleosome complex in ADP(S) state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNP
 
 | | Structure of isw1-nucleosome complex in ATP state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-23 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JO5
 
 | | Structure of isw1-nucleosome complex in ADP-B state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNT
 
 | | Structure of isw1-nucleosome complex in ADP* state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JO2
 
 | | Structure of isw1-nucleosome complex in Apo* state | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNX
 
 | | Structure of isw1-nucleosome complex in ADP*+ state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
