7NQO
 
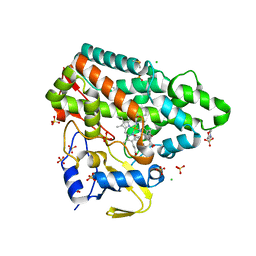 | | Mycobacterium tuberculosis Cytochrome P450 CYP121 in complex with lead compound 21 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[4-[2-(5-bromanyl-1~{H}-indol-3-yl)ethyl]pyrimidin-2-yl]morpholine, CHLORIDE ION, ... | | Authors: | Selvam, I.R. | | Deposit date: | 2021-03-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new strategy for hit generation: Novel in cellulo active inhibitors of CYP121A1 from Mycobacterium tuberculosis via a combined X-ray crystallographic and phenotypic screening approach (XP screen).
Eur.J.Med.Chem., 230, 2022
|
|
7NQM
 
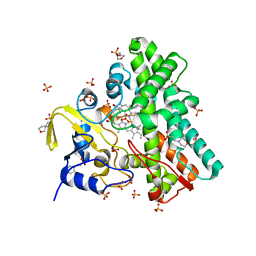 | | Mycobacterium tuberculosis Cytochrome P450 CYP121 in complex with lead compound 10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(2-pyrimidin-4-ylethyl)-1~{H}-indole, CHLORIDE ION, ... | | Authors: | Selvam, I.R. | | Deposit date: | 2021-03-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new strategy for hit generation: Novel in cellulo active inhibitors of CYP121A1 from Mycobacterium tuberculosis via a combined X-ray crystallographic and phenotypic screening approach (XP screen).
Eur.J.Med.Chem., 230, 2022
|
|
7NQN
 
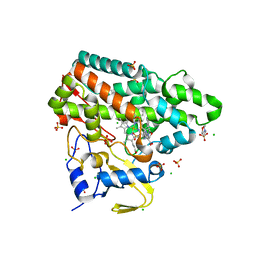 | | Mycobacterium tuberculosis Cytochrome P450 CYP121 in complex with lead compound 14 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[4-[2-(1~{H}-indol-3-yl)ethyl]pyrimidin-2-yl]morpholine, CHLORIDE ION, ... | | Authors: | Selvam, I.R. | | Deposit date: | 2021-03-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new strategy for hit generation: Novel in cellulo active inhibitors of CYP121A1 from Mycobacterium tuberculosis via a combined X-ray crystallographic and phenotypic screening approach (XP screen).
Eur.J.Med.Chem., 230, 2022
|
|
6TL2
 
 | |
4E0W
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | Kainate induces various domain closures in AMPA and kainate receptors.
Neurochem Int, 61, 2012
|
|
4E0X
 
 | |
6YA9
 
 | | Crystal structure of rsGCaMP in the ON state (non-illuminated) | | Descriptor: | CALCIUM ION, rsCGaMP | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-03-11 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 2021
|
|
6APK
 
 | | Trans-acting transferase from Disorazole synthase solved by serial femtosecond XFEL crystallography | | Descriptor: | DisD protein | | Authors: | Lyubimov, A.Y, Mathews, I.I, Uervivojnangkoorn, M, Khosla, C, Soltis, S.M, Cohen, A.E. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
1LOK
 
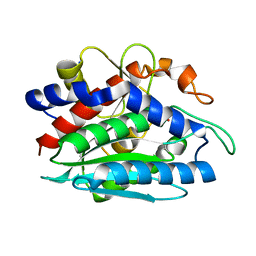 | | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris: A Tale of Buffer Inhibition | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bacterial leucyl aminopeptidase, SODIUM ION, ... | | Authors: | Desmarais, W.T, Bienvenue, D.L, Bzymek, K.P, Holz, R.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris A tale of Buffer Inhibition
Structure, 10, 2002
|
|
6VZK
 
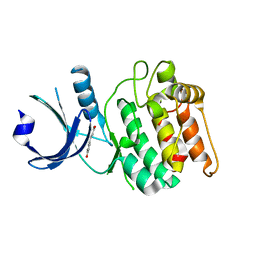 | |
1A38
 
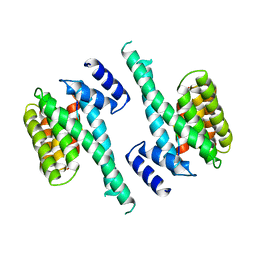 | | 14-3-3 PROTEIN ZETA BOUND TO R18 PEPTIDE | | Descriptor: | 14-3-3 PROTEIN ZETA, R18 PEPTIDE (PHCVPRDLSWLDLEANMCLP) | | Authors: | Petosa, C, Masters, S.C, Pohl, J, Wang, B, Fu, H, Liddington, R.C. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove.
J.Biol.Chem., 273, 1998
|
|
3HCI
 
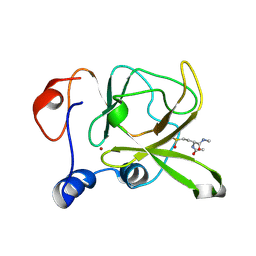 | | Structure of MsrB from Xanthomonas campestris (complex-like form) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, CALCIUM ION, Peptide methionine sulfoxide reductase, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
1NX9
 
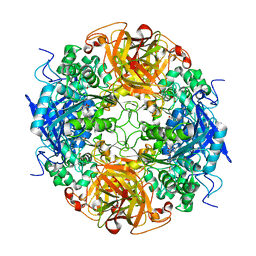 | | Acetobacter turbidans alpha-amino acid ester hydrolase S205A mutant complexed with ampicillin | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, GLYCEROL, alpha-amino acid ester hydrolase | | Authors: | Barends, T.R.M, Polderman-Tijmes, J.J, Jekel, P.A, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-02-10 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acetobacter turbidans {alpha}-Amino Acid Ester Hydrolase: HOW A SINGLE MUTATION IMPROVES AN ANTIBIOTIC-PRODUCING ENZYME.
J.Biol.Chem., 281, 2006
|
|
3HCJ
 
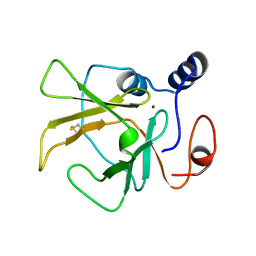 | |
7K2Y
 
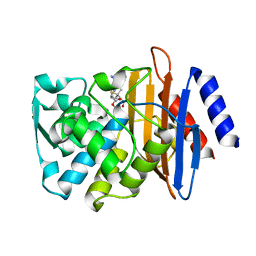 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase | | Authors: | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | Deposit date: | 2020-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
1SJX
 
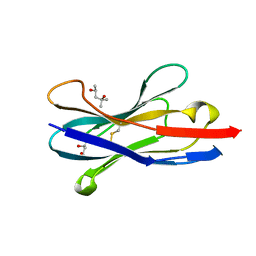 | | Three-Dimensional Structure of a Llama VHH Domain OE7 binding the cell wall protein Malf1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, immunoglobulin VH domain | | Authors: | Dolk, E, van der Vaart, M, Hulsik, D.L, Vriend, G, de Haard, H, Spinelli, S, Cambillau, C, Frenken, L, Verrips, T. | | Deposit date: | 2004-03-04 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isolation of llama antibody fragments for prevention of dandruff by phage display in shampoo.
Appl.Environ.Microbiol., 71, 2005
|
|
6R26
 
 | | The photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | Descriptor: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein, CALCIUM ION | | Authors: | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | Deposit date: | 2019-03-15 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
6R27
 
 | | Crystallographic superstructure of the photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | Descriptor: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | Authors: | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | Deposit date: | 2019-03-15 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
3GS6
 
 | | Vibrio Cholerea family 3 glycoside hydrolase (NagZ)in complex with N-butyryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, GLYCEROL, [[(3R,4R,5S,6R)-3-(BUTANOYLAMINO)-4,5-DIHYDROXY-6-(HYDROXYMETHYL)OXAN-2-YLIDENE]AMINO] N-PHENYLCARBAMATE | | Authors: | Balcewich, M.D, Mark, B.L. | | Deposit date: | 2009-03-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ.
Protein Sci., 18, 2009
|
|
3GSM
 
 | | Vibrio cholerae family 3 glycoside hydrolase (NagZ) bound to N-Valeryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, GLYCEROL, [[(3R,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-(pentanoylamino)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Balcewich, M.D, Mark, B.L. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ.
Protein Sci., 18, 2009
|
|
1MSN
 
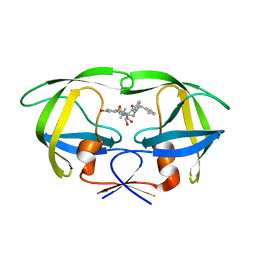 | | The HIV protease (mutant Q7K L33I L63I V82F I84V) complexed with KNI-764 (an inhibitor) | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
1MRX
 
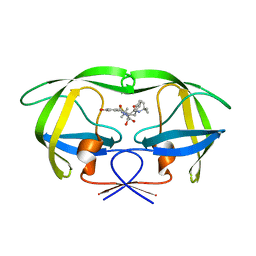 | | Structure of HIV protease (Mutant Q7K L33I L63I V82F I84V ) complexed with KNI-577 | | Descriptor: | (4R)-N-tert-butyl-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-1,3-thiazoli dine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
1OAO
 
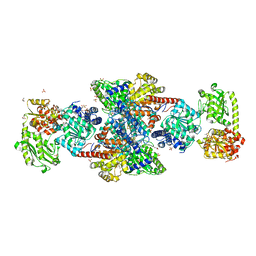 | | NiZn[Fe4S4] and NiNi[Fe4S4] clusters in closed and open alpha subunits of acetyl-CoA synthase/carbon monoxide dehydrogenase | | Descriptor: | ACETATE ION, BICARBONATE ION, CARBON MONOXIDE DEHYDROGENASE/ACETYL-COA SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Darnault, C, Volbeda, A, Kim, E.J, Legrand, P, Vernede, X, Lindahl, P.A, Fontecilla-Camps, J.C. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ni-Zn-[Fe4-S4] and Ni-Ni-[Fe4-S4] Clusters in Closed and Open Alpha Subunits of Acetyl-Coa Synthase/Carbon Monoxide Dehydrogenase
Nat.Struct.Biol., 10, 2003
|
|
7SNL
 
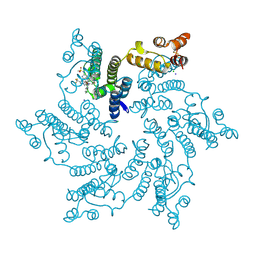 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide | | Descriptor: | CHLORIDE ION, Capsid protein p24, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide
To Be Published
|
|
8GS7
 
 | | SOLUTION NMR STRUCTURE OF N-TERMINAL DOMAIN OF TRICONEPHILA CLAVIPES MAJOR AMPULLATE SPIDROIN 2 | | Descriptor: | Major ampullate spidroin 2 variant 3 | | Authors: | Oktaviani, N.A, Malay, A.D, Matsugami, A, Hayashi, F, Numata, K. | | Deposit date: | 2022-09-05 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unusual p K a Values Mediate the Self-Assembly of Spider Dragline Silk Proteins.
Biomacromolecules, 24, 2023
|
|
