8GOO
 
 | | Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 | | Descriptor: | Beta-arrestin-2, C5a anaphylatoxin chemotactic receptor 1, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
6H6P
 
 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, Ubiquinone biosynthesis protein UbiJ | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
6RXY
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state a | | Descriptor: | 35S rRNA, 40S ribosomal protein S13-like protein, 40S ribosomal protein S14-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6HCJ
 
 | | Structure of the rabbit 80S ribosome on globin mRNA in the rotated state with A/P and P/E tRNAs | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6RXT
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state A | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
8GVL
 
 | | PTPN21 FERM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8GWH
 
 | |
6RXZ
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state b | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S11-like protein, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
8GVV
 
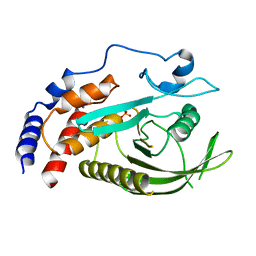 | | PTPN21 PTP domain C1108S mutant | | Descriptor: | IODIDE ION, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8GXE
 
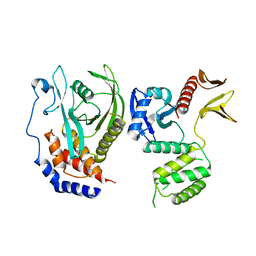 | | PTPN21 FERM PTP complex | | Descriptor: | CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
6RXX
 
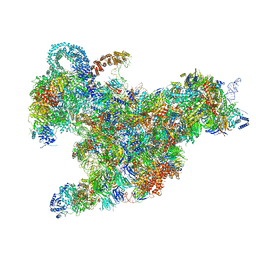 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state C, Poly-Ala | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6S6B
 
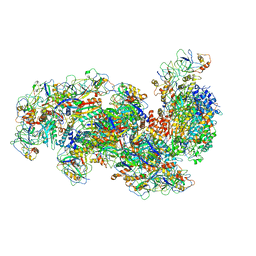 | | Type III-B Cmr-beta Cryo-EM structure of the Apo state | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6HCQ
 
 | | Structure of the rabbit collided di-ribosome (collided monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCM
 
 | | Structure of the rabbit collided di-ribosome (stalled monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HHO
 
 | | Crystal structure of RIP1 kinase in complex with GSK547 | | Descriptor: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
6RXV
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B2 | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6J9M
 
 | | NmeBH+AcrIIC2 | | Descriptor: | AcrIIC2, CRISPR-associated endonuclease Cas9 | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6J9K
 
 | | Apo-AcrIIC2 | | Descriptor: | AcrIIC2, MAGNESIUM ION | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6S8E
 
 | | Cryo-EM structure of the type III-B Cmr-beta complex bound to non-cognate target RNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S91
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2 | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6QH3
 
 | |
6J9L
 
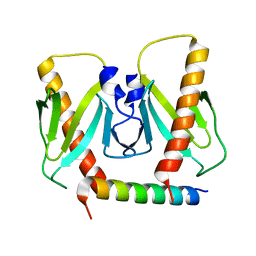 | | FnoBH+AcrIIC2 | | Descriptor: | AcrIIC2, HNH endonuclease family protein | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6SIC
 
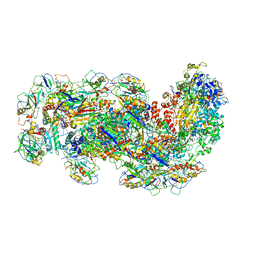 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr1 family, Cmr4 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6R1T
 
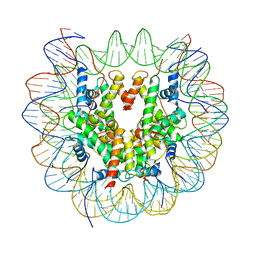 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | Descriptor: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R1U
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 | | Descriptor: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
