3QLC
 
 | |
3QO2
 
 | | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9 | | Descriptor: | 1,2-ETHANEDIOL, Histone H3 peptide, M-phase phosphoprotein 8 | | Authors: | Chang, Y, Horton, J.R, Bedford, M.T, Zhang, X, Cheng, X. | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding.
J.Mol.Biol., 408, 2011
|
|
2RSN
 
 | |
6VFO
 
 | | Solution structure of the PHD of mouse UHRF1 (NP95) | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Lemak, A, Houliston, S, Duan, S, Arrowsmith, C.H. | | Deposit date: | 2020-01-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alternative splicing and allosteric regulation modulate the chromatin binding of UHRF1.
Nucleic Acids Res., 48, 2020
|
|
6VEE
 
 | |
6VED
 
 | | Solution structure of the TTD and linker region of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1 | | Authors: | Lemak, A, Houliston, S, Duan, S, Ong, M.S, Arrowsmith, C.H. | | Deposit date: | 2019-12-31 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alternative splicing and allosteric regulation modulate the chromatin binding of UHRF1.
Nucleic Acids Res., 48, 2020
|
|
7M5U
 
 | | Crystal structure of human MPP8 chromodomain in complex with peptidomimetic ligand UNC5246 | | Descriptor: | M-phase phosphoprotein 8, UNC5246 | | Authors: | Budziszewski, G.R, McGinty, R.K, Waybright, J.M, Norris, J.L, James, L.I. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A Peptidomimetic Ligand Targeting the Chromodomain of MPP8 Reveals HRP2's Association with the HUSH Complex.
Acs Chem.Biol., 16, 2021
|
|
4IUR
 
 | |
6SWG
 
 | |
6MHA
 
 | | dHP1 Chromodomain Y24W variant bound to histone H3 peptide containing trimethyllysine | | Descriptor: | Heterochromatin protein 1, histone H3 peptide containing trimethyllysine, H3K9me3 | | Authors: | Brustad, E.M, Krone, M.W, Albanese, K.I, Waters, M.L. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Thermodynamic consequences of Tyr to Trp mutations in the cation-pi-mediated binding of trimethyllysine by the HP1 chromodomain
Chem Sci, 11, 2020
|
|
8QFB
 
 | |
6V2S
 
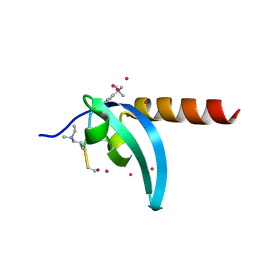 | | Crystal Structure of chromodomain of MPP8 in complex with inhibitor UNC3866 | | Descriptor: | M-phase phosphoprotein 8, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6D07
 
 | |
7EF2
 
 | |
3G7L
 
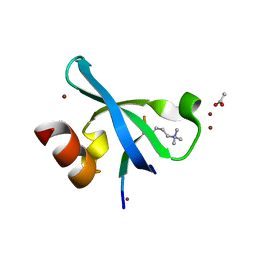 | | Chromodomain of Chp1 in complex with Histone H3K9me3 peptide | | Descriptor: | ACETIC ACID, Chromo domain-containing protein 1, Histone H3.1/H3.2, ... | | Authors: | Schalch, T, Joshua-Tor, L. | | Deposit date: | 2009-02-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin
Mol.Cell, 34, 2009
|
|
2RVN
 
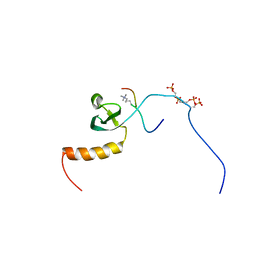 | |
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
4HOO
 
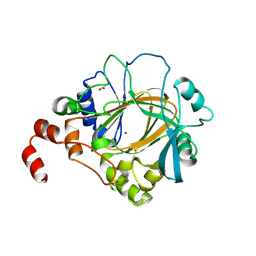 | | Crystal structure of human JMJD2D/KDM4D apoenzyme | | Descriptor: | ACETATE ION, Lysine-specific demethylase 4D, NICKEL (II) ION, ... | | Authors: | Krishnan, S, Trievel, R.C. | | Deposit date: | 2012-10-22 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structural and Functional Analysis of JMJD2D Reveals Molecular Basis for Site-Specific Demethylation among JMJD2 Demethylases.
Structure, 21, 2013
|
|
9BAP
 
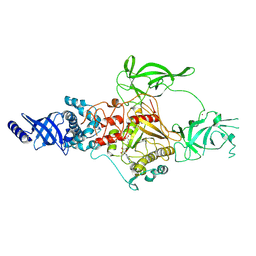 | | CryoEM structure of Apo-DIM2 | | Descriptor: | DNA (cytosine-5-)-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
9BAZ
 
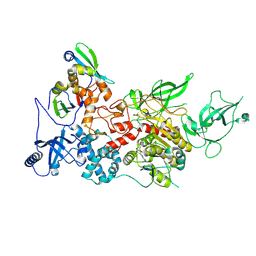 | | CryoEM structure of DIM2-HP1 complex | | Descriptor: | DNA (cytosine-5-)-methyltransferase, Heterochromatin protein one, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-05 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
7BU9
 
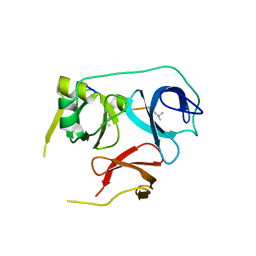 | | Crystal Structure of Spindlin1-H3(K4me3-K9me2) complex | | Descriptor: | H3(K4me3-K9me2) peptide, Spindlin-1 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2020-04-05 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Molecular basis for histone H3 "K4me3-K9me3/2" methylation pattern readout by Spindlin1.
J.Biol.Chem., 295, 2020
|
|
7BQZ
 
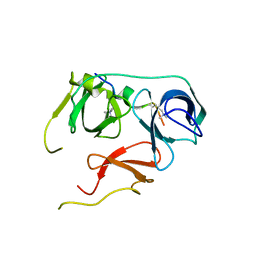 | |
3AVR
 
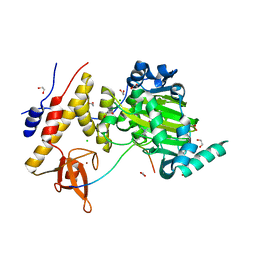 | | Catalytic fragment of UTX/KDM6A bound with histone H3K27me3 peptide, N-oxyalylglycine, and Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone H3, ... | | Authors: | Sengoku, T, Yokoyama, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
3AVS
 
 | | Catalytic fragment of UTX/KDM6A bound with N-oxyalylglycine, and Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 6A, ... | | Authors: | Sengoku, T, Yokoyama, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
3U5M
 
 | |
