5VHW
 
 | | GluA2-0xGSG1L bound to ZK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Germ cell-specific gene 1-like protein, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes.
Neuron, 94, 2017
|
|
2BQV
 
 | | HIV-1 protease in complex with inhibitor AHA455 | | Descriptor: | 6-AMINO HEXANOIC ACID, HIV-1 PROTEASE | | Authors: | Unge, T, Ekegren, J.K, Schenk, H.V, Zreik Safa, M, Wallberg, H, Samuelsson, B, Hallberg, A. | | Deposit date: | 2005-04-28 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A New Class of HIV-1 Protease Inhibitors Containing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 48, 2005
|
|
7ZU9
 
 | |
5VQD
 
 | | Beta-glucoside phosphorylase BglX | | Descriptor: | Beta-glucoside phosphorylase BglX, GLYCEROL | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
6ZYT
 
 | | Monomeric streptavidin with a conjugated biotinylated pyrrolidine | | Descriptor: | 5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-N-((S)-pyrrolidin-3-yl)pentanamide, SULFATE ION, Streptavidin/Rhizavidin Hybrid | | Authors: | Nodling, A.R, Lipka-Lloyd, M, Tsai, Y.H, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of streptavidin and its variants in catalysis by biotinylated secondary amines.
Org.Biomol.Chem., 19, 2021
|
|
5OLS
 
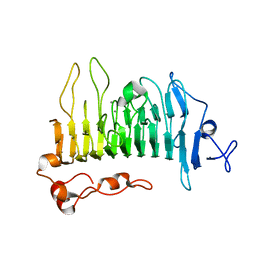 | | Rhamnogalacturonan lyase | | Descriptor: | 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-2)-alpha-L-rhamnopyranose-(1-4)-beta-D-galactopyranuronic acid, CALCIUM ION, Rhamnogalacturonan lyase, ... | | Authors: | Basle, A, Luis, A.S, Gilbert, H.J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
5VHZ
 
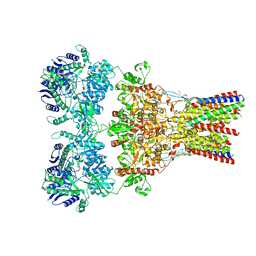 | | GluA2-2xGSG1L bound to L-Quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2,Germ cell-specific gene 1-like protein | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-03 | | Last modified: | 2017-11-08 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes.
Neuron, 94, 2017
|
|
5ILP
 
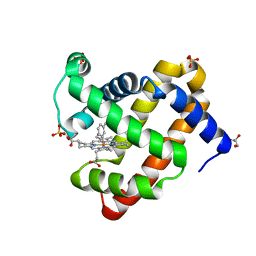 | | H64Q sperm whale myoglobin with a Fe-tolyl moiety | | Descriptor: | GLYCEROL, Myoglobin, SULFATE ION, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2016-03-04 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Organometallic myoglobins: Formation of Fe-carbon bonds and distal pocket effects on aryl ligand conformations.
J. Inorg. Biochem., 164, 2016
|
|
5US3
 
 | |
8IL6
 
 | |
8Y9P
 
 | |
5FPS
 
 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 3-aminobenzene-1,2-dicarboxylic acid (AT1246) in an alternate binding site. | | Descriptor: | 3-AMINOBENZENE-1,2-DICARBOXYLIC ACID, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8TMA
 
 | |
7SG4
 
 | |
8TM1
 
 | |
8JTJ
 
 | | Cryo-EM structure of GeoCas9-sgRNA-dsDNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (29-MER), DNA (5'-D(P*GP*GP*GP*CP*GP*CP*GP*AP*A)-3'), ... | | Authors: | Shen, P.P, Liu, B.B, Li, X, Zhang, L.L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structure of GeoCas9-sgRNA-DNA ternary complex
To Be Published
|
|
8IL2
 
 | |
5D0K
 
 | | Structure of UbE2D2:RNF165:Ub complex | | Descriptor: | Polyubiquitin-B, RING finger protein 165, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Wright, J.D, Day, C.L, Mace, P.D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5FTI
 
 | |
8A9M
 
 | | Hippeastrum hybrid agglutinin, HHA, complex with beta-mannose | | Descriptor: | Agglutinin, PHOSPHATE ION, beta-D-mannopyranose | | Authors: | Rizkallah, P.J. | | Deposit date: | 2022-06-28 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure solution of amaryllis lectin by molecular replacement with only 4% of the total diffracting matter.
Acta Crystallogr D Biol Crystallogr, 52, 1996
|
|
5D0I
 
 | | Structure of RING finger protein 165 | | Descriptor: | RING finger protein 165, SULFATE ION, ZINC ION | | Authors: | Wright, J.D, Day, C.L, Mace, P.D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D0M
 
 | | Structure of UbE2D2:RNF165:Ub complex | | Descriptor: | PHOSPHATE ION, Polyubiquitin-B, RING finger protein 165, ... | | Authors: | Wright, J.D, Day, C.L, Mace, P.D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-12-09 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (1.913 Å) | | Cite: | Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5Y60
 
 | | V/A-type ATPase/synthase from Thermus thermophilus, rotational state 3. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nat Commun, 9, 2018
|
|
5IKS
 
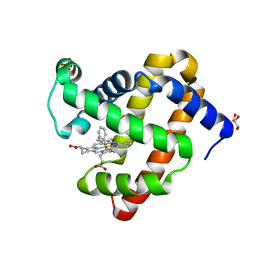 | | Wild-type sperm whale myoglobin with a Fe-phenyl moiety | | Descriptor: | GLYCEROL, Myoglobin, SULFATE ION, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2016-03-03 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Organometallic myoglobins: Formation of Fe-carbon bonds and distal pocket effects on aryl ligand conformations.
J. Inorg. Biochem., 164, 2016
|
|
2B5I
 
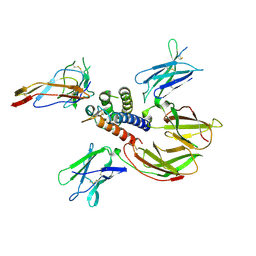 | | cytokine receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common gamma chain, ... | | Authors: | Wang, X, Rickert, M, Garcia, K.C. | | Deposit date: | 2005-09-28 | | Release date: | 2005-11-29 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors.
Science, 310, 2005
|
|
