9GEW
 
 | | Crystal structure of CREBBP bromodomain in complex with (R,E)-6-(5-(6-methoxy-2,3-dihydro-4H-benzo[b][1,4]oxazin-4-yl)pent-1-en-1-yl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one | | Descriptor: | (4~{R})-6-[(~{E})-5-(6-methoxy-2,3-dihydro-1,4-benzoxazin-4-yl)pent-1-enyl]-4-methyl-1,3,4,5-tetrahydro-1,5-benzodiazepin-2-one, (R,R)-2,3-BUTANEDIOL, CREBBP | | Authors: | Amann, M, Boyd, A, Einsle, O, Moroglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure of CREBBP bromodomain in complex with (R,E)-6-(5-(6-methoxy-2,3-dihydro-4H-benzo[b][1,4]oxazin-4-yl)pent-1-en-1-yl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one
To Be Published
|
|
9GEJ
 
 | | Crystal structure of CREBBP bromodomain in complex with (2R,13S,E)-2-methyl-1,2,3,5,10,11,13,14,20,21,24,25-dodecahydro-19H,23H-16,18-etheno-9,13-methano-7,28-(metheno)[1,4]diazepino[2,3-k]pyrido[1,2-s][1,4]dioxa[7,19]diazacyclodocosine-4,8-dione | | Descriptor: | 2R,13S,E)-2-methyl-1,2,3,5,10,11,13,14,20,21,24,25-dodecahydro-19H,23H-16,18-etheno-9,13-methano-7,28-(metheno)[1,4]diazepino[2,3-k]pyrido[1,2-s][1,4]dioxa[7,19]diazacyclodocosine-4,8-dione, CREBBP, SULFATE ION | | Authors: | Amann, M, Boyd, A, Einsle, O, Guenther, S, Moroglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of CREBBP bromodomain in complex with (2R,13S,E)-2-methyl-1,2,3,5,10,11,13,14,20,21,24,25-dodecahydro-19H,23H-16,18-etheno-9,13-methano-7,28-(metheno)[1,4]diazepino[2,3-k]pyrido[1,2-s][1,4]dioxa[7,19]diazacyclodocosine-4,8-dione
To Be Published
|
|
9GEY
 
 | | Crystal structure of CREBBP bromodomain in complex with (R,E)-2-methyl-1,2,3,5,9,10,11,12,18,19,22,23-dodecahydro-17H,21H-14,16-etheno-7,26-(metheno)[1,4]diazepino[2,3-l]pyrido[2,1-d][1]oxa[5,17]diazacycloicosine-4,8-dione | | Descriptor: | (R,E)-2-methyl-1,2,3,5,9,10,11,12,18,19,22,23-dodecahydro-17H,21H-14,16-etheno-7,26-(metheno)[1,4]diazepino[2,3-l]pyrido[2,1-d][1]oxa[5,17]diazacycloicosine-4,8-dione, 1,2-ETHANEDIOL, CREBBP | | Authors: | Amann, M, Boyd, A, Einsle, O, Guenther, S, Moroglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of CREBBP bromodomain in complex with (R,E)-2-methyl-1,2,3,5,9,10,11,12,18,19,22,23-dodecahydro-17H,21H-14,16-etheno-7,26-(metheno)[1,4]diazepino[2,3-l]pyrido[2,1-d][1]oxa[5,17]diazacycloicosine-4,8-dione
To Be Published
|
|
9GEU
 
 | | Crystal structure of CREBBP bromodomain in complex with (R,E)-2-methyl-1,2,3,5,10,11,17,18,21,22-decahydro-4H,16H,20H-13,15-etheno-7,25-(metheno)[1,4]diazepino[2,3-h]pyrido[1,2-p][1]oxa[4,16]diazacyclononadecine-4,8(9H)-dione | | Descriptor: | (R,E)-2-methyl-1,2,3,5,10,11,17,18,21,22-decahydro-4H,16H,20H-13,15-etheno-7,25-(metheno)[1,4]diazepino[2,3-h]pyrido[1,2-p][1]oxa[4,16]diazacyclononadecine-4,8(9H)-dione, (R,R)-2,3-BUTANEDIOL, CITRIC ACID, ... | | Authors: | Amann, M, Boyd, A, Einsle, O, Guenther, S, Moroglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the human CREBBP bromodomain in complex with (R,E)-2-methyl-1,2,3,5,10,11,17,18,21,22-decahydro-4H,16H,20H-13,15-etheno-7,25-(metheno)[1,4]diazepino[2,3-h]pyrido[1,2-p][1]oxa[4,16]diazacyclononadecine-4,8(9H)-dione
To Be Published
|
|
6NIJ
 
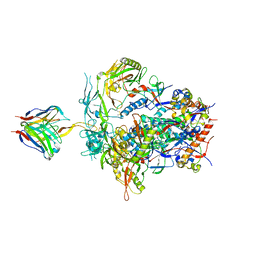 | | PGT145 Fab in complex with full length AMC011 HIV-1 Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC011 Glycoprotein 120, ... | | Authors: | Cottrell, C.A, Torrents de la Pena, A, Rantalainen, K, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-29 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics.
Plos Pathog., 15, 2019
|
|
6NBQ
 
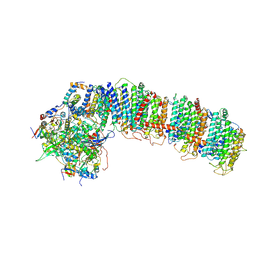 | | T.elongatus NDH (data-set 1) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 2, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
8Q6L
 
 | | human Carbonic Anhydrase I in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol | | Descriptor: | 1,1-bis(oxidanyl)-3,4-dihydro-2,1$l^{4}-benzoxaborinine, Carbonic anhydrase 1, SODIUM ION, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-08-13 | | Release date: | 2024-08-21 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Benzoxaborinine, New Chemotype for Carbonic Anhydrase Inhibition: Ex Novo Synthesis, Crystallography, In Silico Studies, and Anti-Melanoma Cell Line Activity.
J.Med.Chem., 67, 2024
|
|
8Q7G
 
 | | Human Carbonic Anhydrase I in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol pH 7.0 | | Descriptor: | 1,1-bis(oxidanyl)-3,4-dihydro-2,1$l^{4}-benzoxaborinine, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-28 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Benzoxaborinine, New Chemotype for Carbonic Anhydrase Inhibition: Ex Novo Synthesis, Crystallography, In Silico Studies, and Anti-Melanoma Cell Line Activity.
J.Med.Chem., 67, 2024
|
|
6VZI
 
 | |
8CX1
 
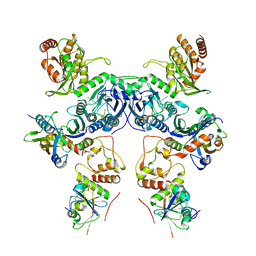 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
3EU8
 
 | |
6X9T
 
 | |
6X5U
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and NM5M2-Asn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
7BG7
 
 | | HRV14 in complex with its receptor ICAM-1 | | Descriptor: | Genome polyprotein, Intercellular adhesion molecule 1 | | Authors: | Hrebik, D, Fuzik, T, Plevka, P. | | Deposit date: | 2021-01-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NKG
 
 | |
8CRM
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11C6 refined against anomalous diffraction data | | Descriptor: | 1-[2-(3-chlorophenyl)-1,3-thiazol-4-yl]-~{N}-methyl-methanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
9D7A
 
 | | OXA-58-NA-1-157 5 min complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase | | Authors: | Smith, C.A, Maggiolo, A.O, Toth, M, Vakulenko, S.B. | | Deposit date: | 2024-08-16 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Decarboxylation of the Catalytic Lysine Residue by the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the OXA-58 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
9D7C
 
 | | OXA-58-NA-1-157 10 min complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CARBON DIOXIDE | | Authors: | Smith, C.A, Maggiolo, A.O, Toth, M, Vakulenko, S.B. | | Deposit date: | 2024-08-16 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Decarboxylation of the Catalytic Lysine Residue by the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the OXA-58 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
7BJX
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 26 | | Descriptor: | 4-amino-7-methyl-2-[(1-methyl-1H-pyrazol-4-yl)amino]-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BK3
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 45 | | Descriptor: | 4-amino-7-methyl-2-({5-methyl-1-[(3S)-oxolan-3-yl]-1H-pyrazol-4-yl}amino)-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BK2
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 44 | | Descriptor: | 4-amino-7-methyl-2-({5-methyl-1-[(3R)-oxolan-3-yl]-1H-pyrazol-4-yl}amino)-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BK1
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 32 | | Descriptor: | 4-amino-2-[(1,3-dimethyl-1H-pyrazol-4-yl)amino]-7-methyl-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
6NX1
 
 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN BOUND TETHERED WITH SRC CO-ACTIVATOR PEPTIDE AND COMPOUND-3 AKA 1,1,1,3,3,3-HEXAFLUORO-2-{4-[1-(4- LUOROBENZENESULFONYL)CYCLOPENTYL]PHENYL}PROPAN-2-OL | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-(4-{1-[(4-fluorophenyl)sulfonyl]cyclopentyl}phenyl)propan-2-ol, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 fusion | | Authors: | Khan, J.A. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-based Discovery of Phenyl (3-Phenylpyrrolidin-3-yl)sulfones as Selective, Orally Active ROR gamma t Inverse Agonists.
Acs Med.Chem.Lett., 10, 2019
|
|
6RTJ
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1-[(dimethylamino)methyl]-2-naphthol at 1 hour of soaking | | Descriptor: | 1-[(dimethylamino)methyl]naphthalen-2-ol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Angelucci, F, Silvestri, I, Fata, F, Williams, D.L. | | Deposit date: | 2019-05-24 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ectopic suicide inhibition of thioredoxin glutathione reductase.
Free Radic Biol Med, 147, 2020
|
|
5N2K
 
 | | Structure of unbound Briakinumab FAb | | Descriptor: | 1,2-ETHANEDIOL, Briakinumab FAb heavy chain, Briakinumab FAb light chain | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2017-02-07 | | Release date: | 2018-01-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.219 Å) | | Cite: | Structural Activation of Pro-inflammatory Human Cytokine IL-23 by Cognate IL-23 Receptor Enables Recruitment of the Shared Receptor IL-12R beta 1.
Immunity, 48, 2018
|
|
