3PZP
 
 | | Human DNA polymerase kappa extending opposite a cis-syn thymine dimer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*CP*A)-3', 5'-D(*TP*TP*CP*CP*(TTD)P*GP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Vasquez-Del Carpio, R, Silverstein, T.D, Lone, S, Johnson, R.E, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.336 Å) | | Cite: | Role of human DNA polymerase kappa in extension opposite from a cis-syn thymine dimer.
J.Mol.Biol., 408, 2011
|
|
4EEY
 
 | | Crystal structure of human DNA polymerase eta in ternary complex with a cisplatin DNA adduct | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*TP*GP*GP*TP*CP*TP*CP*CP*TP*CP*C)-3', 5'-D(*TP*GP*GP*AP*GP*GP*AP*GP*A)-3', ... | | Authors: | Ummat, A, Rechkoblit, O, Jain, R, Choudhury, J.R, Johnson, R.E, Silverstein, T.D, Buku, A, Lone, S, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for cisplatin DNA damage tolerance by human polymerase {eta} during cancer chemotherapy.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3Q8P
 
 | | Human DNA polymerase iota incorporating dCTP opposite 8-oxo-guanine | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*TP*CP*AP*(8OG)P*GP*GP*GP*TP*CP*CP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Kirouac, K.N, Ling, H. | | Deposit date: | 2011-01-06 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase iota.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3Q8Q
 
 | | Human DNA polymerase iota incorporating dATP opposite 8-oxo-guanine | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*TP*CP*AP*(8OG)P*GP*GP*GP*TP*CP*CP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Kirouac, K.N, Ling, H. | | Deposit date: | 2011-01-06 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase iota.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6I1L
 
 | |
3IFZ
 
 | | crystal structure of the first part of the Mycobacterium tuberculosis DNA gyrase reaction core: the breakage and reunion domain at 2.7 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit A, SODIUM ION | | Authors: | Piton, J, Aubry, A, Delarue, M, Mayer, C. | | Deposit date: | 2009-07-27 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase.
Plos One, 5, 2010
|
|
3Q8R
 
 | | Human DNA polymerase iota incorporating dGTP opposite 8-oxo-guanine | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*TP*CP*AP*(8OG)P*GP*GP*GP*TP*CP*CP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Kirouac, K.N, Ling, H. | | Deposit date: | 2011-01-06 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase iota.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3WAZ
 
 | | Crystal structure of a restriction enzyme PabI in complex with DNA | | Descriptor: | ADENINE, DNA (5'-D(*GP*CP*AP*TP*AP*GP*CP*TP*GP*TP*(ORP)P*CP*AP*GP*CP*TP*AP*TP*GP*C)-3'), Putative uncharacterized protein | | Authors: | Miyazono, K, Miyakawa, T, Ito, T, Tanokura, M. | | Deposit date: | 2013-05-10 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A sequence-specific DNA glycosylase mediates restriction-modification in Pyrococcus abyssi.
Nat Commun, 5, 2014
|
|
8QAM
 
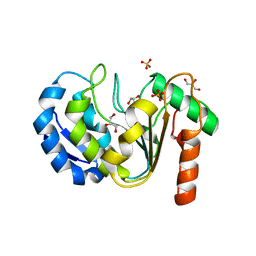 | |
5CDM
 
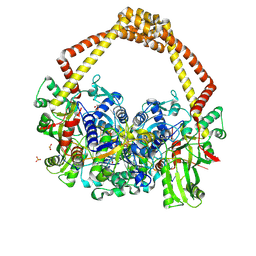 | | 2.5A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4',6'(1'H,3'H)-trione, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
3OSP
 
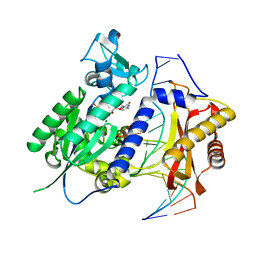 | | Structure of rev1 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', 5'-D(*TP*AP*AP*(3DR)P*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*AP*T)-3', ... | | Authors: | Nair, D.T, Aggarwal, A.K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA synthesis across an abasic lesion by yeast REV1 DNA polymerase.
J.Mol.Biol., 406, 2011
|
|
1MWI
 
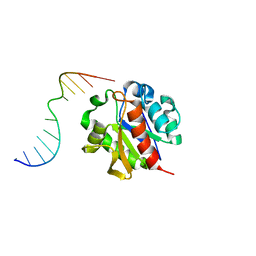 | | Crystal structure of a MUG-DNA product complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*GP*(AAB)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | Authors: | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | Deposit date: | 2002-09-30 | | Release date: | 2002-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
7LMK
 
 | |
4B21
 
 | | Unprecedented sculpting of DNA at abasic sites by DNA glycosylase homolog Mag2 | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP)-3', 5'-D(*GP*CP*TP*AP*CP*3DRP*GP*AP*TP*CP*GP)-3', PHOSPHATE ION, ... | | Authors: | Dalhus, B, Nilsen, L, Korvald, H, Huffman, J, Forstrom, R.J, McMurray, C.T, Alseth, I, Tainer, J.A, Bjoras, M. | | Deposit date: | 2012-07-12 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Sculpting of DNA at Abasic Sites by DNA Glycosylase Homolog Mag2.
Structure, 21, 2013
|
|
8BP2
 
 | | 2.8A STRUCTURE OF ZOLIFLODACIN WITH S.AUREUS DNA GYRASE AND DNA | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (20-MER), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Morgan, H, Warren, A.J. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A 2.8 angstrom Structure of Zoliflodacin in a DNA Cleavage Complex with Staphylococcus aureus DNA Gyrase.
Int J Mol Sci, 24, 2023
|
|
6TYW
 
 | |
6TYZ
 
 | |
6TYV
 
 | |
6TYX
 
 | |
7W5P
 
 | | Crystal Structure of the dioxygenase CcTet from Coprinopsis cinereain bound to 12bp N6-methyldeoxyadenine (6mA) containing duplex DNA | | Descriptor: | CcTet, DNA, DNA (12-MER), ... | | Authors: | Mu, Y.J, Zhang, L, Zhang, L. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A fungal dioxygenase CcTet serves as a eukaryotic 6mA demethylase on duplex DNA.
Nat.Chem.Biol., 18, 2022
|
|
7YHS
 
 | | Structure of Csy-AcrIF4-dsDNA | | Descriptor: | AcrIF4, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Feng, Y, Zhang, L.X. | | Deposit date: | 2022-07-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Anti-CRISPR protein AcrIF4 inhibits the type I-F CRISPR-Cas surveillance complex by blocking nuclease recruitment and DNA cleavage.
J.Biol.Chem., 298, 2022
|
|
6TYU
 
 | |
4B24
 
 | | Unprecedented sculpting of DNA at abasic sites by DNA glycosylase homolog Mag2 | | Descriptor: | 5'-D(*CP*GP*AP*TP*AP*GP*GP*TP*AP*GP)-3', 5'-D(*GP*CP*TP*AP*CP*3DRP*TP*AP*TP*CP*GP)-3', PROBABLE DNA-3-METHYLADENINE GLYCOSYLASE 2 | | Authors: | Dalhus, B, Nilsen, L, Korvald, H, Huffman, J, Forstrom, R.J, McMurray, C.T, Alseth, I, Tainer, J.A, Bjoras, M. | | Deposit date: | 2012-07-12 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sculpting of DNA at Abasic Sites by DNA Glycosylase Homolog Mag2.
Structure, 21, 2013
|
|
7VW3
 
 | | Cryo-EM structure of SaCas9-sgRNA-DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, MAGNESIUM ION, Non-target DNA strand, ... | | Authors: | Du, W.H, Qian, J.Q, Huang, Q. | | Deposit date: | 2021-11-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Full-Length Model of SaCas9-sgRNA-DNA Complex in Cleavage State.
Int J Mol Sci, 24, 2023
|
|
6TYT
 
 | |
