8BU9
 
 | | Structure of DDB1 bound to roscovitine-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUP
 
 | | Structure of DDB1 bound to DS30-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-[3-(3-methylphenyl)propylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUB
 
 | | Structure of DDB1 bound to dCeMM4-engaged CDK12-cyclin K | | Descriptor: | CITRIC ACID, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUL
 
 | | Structure of DDB1 bound to DS11-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-(3-phenylpropylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, 1,2-ETHANEDIOL, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU3
 
 | | Structure of DDB1 bound to DS19-engaged CDK12-cyclin K | | Descriptor: | 1,2-ETHANEDIOL, 2-morpholin-4-yl-9-propan-2-yl-~{N}-[(4-pyridin-2-ylphenyl)methyl]purin-6-amine, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUQ
 
 | | Structure of DDB1 bound to DS43-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-[[1-(3-chlorophenyl)pyrazol-3-yl]methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, CITRIC ACID, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUC
 
 | | Structure of DDB1 bound to dCeMM3-engaged CDK12-cyclin K | | Descriptor: | 2-(1~{H}-benzimidazol-2-ylsulfanyl)-~{N}-(5-chloranylpyridin-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU5
 
 | | Structure of DDB1 bound to SR-4835-engaged CDK12-cyclin K | | Descriptor: | CITRIC ACID, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.134 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUT
 
 | | Structure of DDB1 bound to DS61-engaged CDK12-cyclin K | | Descriptor: | 2-[[6-[[4-(2-hydroxyethyloxy)phenyl]methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUS
 
 | | Structure of DDB1 bound to DS59-engaged CDK12-cyclin K | | Descriptor: | 1,3-dimethyl-5-[[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)methylamino]purin-2-yl]amino]methyl]pyrazole-4-sulfonamide, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUH
 
 | | Structure of DDB1 bound to WX3-engaged CDK12-cyclin K | | Descriptor: | 6-[[[2-[[(2~{R})-1-oxidanylbutan-2-yl]amino]-9-propan-2-yl-purin-6-yl]amino]methyl]-3-pyridin-2-yl-1~{H}-pyridin-2-one, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUD
 
 | | Structure of DDB1 bound to Z7-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUE
 
 | | Structure of DDB1 bound to Z11-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUG
 
 | | Structure of DDB1 bound to HQ461-engaged CDK12-cyclin K | | Descriptor: | 2-[2-[(6-methylpyridin-2-yl)amino]-1,3-thiazol-4-yl]-~{N}-(5-methyl-1,3-thiazol-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
3LJ2
 
 | | IRE1 complexed with JAK Inhibitor I | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Lee, K.P.K, Sicheri, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1.
Mol.Cell, 38, 2010
|
|
5EFQ
 
 | | Crystal structure of human Cdk13/Cyclin K in complex with ADP-aluminum fluoride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin-K, ... | | Authors: | Hoenig, D, Greifenberg, A.K, Anand, K, Geyer, M. | | Deposit date: | 2015-10-24 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of the Cdk13/Cyclin K Complex.
Cell Rep, 14, 2016
|
|
3MWU
 
 | | Activated Calcium-Dependent Protein Kinase 1 from Cryptosporidium parvum (CpCDPK1) in complex with bumped kinase inhibitor RM-1-95 | | Descriptor: | 3-(naphthalen-1-ylmethyl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, Calmodulin-domain protein kinase 1 | | Authors: | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2010-05-06 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Calcium-Dependent Protein Kinase 1 (CDPK1) from C. parvum and T. gondii.
ACS Med Chem Lett, 1, 2010
|
|
3MVH
 
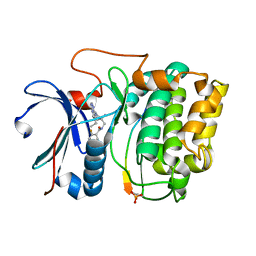 | | Crystal structure of Akt-1-inhibitor complexes | | Descriptor: | GSK3-beta peptide, MANGANESE (II) ION, N-{[(3S)-3-amino-1-(5-ethyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-yl]methyl}-2,4-difluorobenzamide, ... | | Authors: | Pandit, J. | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-02 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
5CZO
 
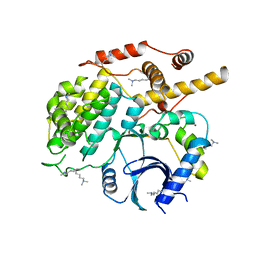 | | Structure of S. cerevisiae Hrr25:Mam1 complex, form 2 | | Descriptor: | Casein kinase I homolog HRR25, Monopolin complex subunit MAM1, ZINC ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
3KRW
 
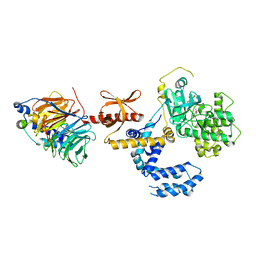 | | Human GRK2 in complex with Gbetgamma subunits and balanol (soak) | | Descriptor: | BALANOL, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tesmer, J.J.G, Tesmer, V.M. | | Deposit date: | 2009-11-19 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human g protein-coupled receptor kinase 2 in complex with the kinase inhibitor balanol.
J.Med.Chem., 53, 2010
|
|
3MY1
 
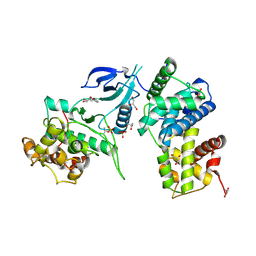 | | Structure of CDK9/cyclinT1 in complex with DRB | | Descriptor: | 5,6-dichloro-1-beta-D-ribofuranosyl-1H-benzimidazole, Cell division protein kinase 9, Cyclin-T1, ... | | Authors: | Baumli, S, Johnson, L.N. | | Deposit date: | 2010-05-09 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Halogen bonds form the basis for selective P-TEFb inhibition by DRB
Chem.Biol., 17, 2010
|
|
3N51
 
 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with bumped kinase inhibitor RM-1-95 | | Descriptor: | 1,2-ETHANEDIOL, 3-(naphthalen-1-ylmethyl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, ... | | Authors: | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Calcium-Dependent Protein Kinase 1 (CDPK1) from C. parvum and T. gondii.
ACS Med Chem Lett, 1, 2010
|
|
3MY5
 
 | | CDk2/cyclinA in complex with DRB | | Descriptor: | 5,6-dichloro-1-beta-D-ribofuranosyl-1H-benzimidazole, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Baumli, S, Johnson, L.N. | | Deposit date: | 2010-05-09 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Halogen bonds form the basis for selective P-TEFb inhibition by DRB
Chem.Biol., 17, 2010
|
|
5ES1
 
 | |
8BW9
 
 | | Cryo-EM structure of the RAF activating complex KSR-MEK-CNK-HYP | | Descriptor: | Connector enhancer of KSR protein CNK, Dual specificity mitogen-activated protein kinase kinase dSOR1, KSR, ... | | Authors: | Maisonneuve, P, Fronzes, R, Sicheri, F. | | Deposit date: | 2022-12-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | The CNK-HYP scaffolding complex promotes RAF activation by enhancing KSR-MEK interaction.
Nat.Struct.Mol.Biol., 31, 2024
|
|
