5X41
 
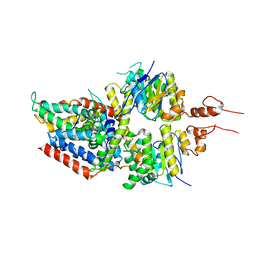 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
3L02
 
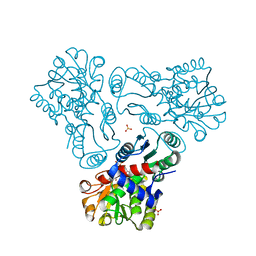 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase E92A mutant complexed with carbamyl phosphate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
Protein Sci., 16, 2007
|
|
3H6S
 
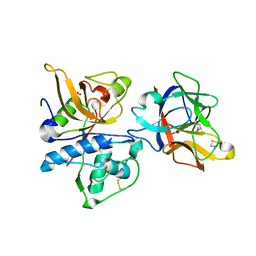 | | Structure of clitocypin - cathepsin V complex | | Descriptor: | Cathepsin L2, Clitocypin analog, SULFATE ION | | Authors: | Renko, M, Sabotic, J, Brzin, J, Turk, D. | | Deposit date: | 2009-04-23 | | Release date: | 2009-10-20 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Versatile loops in mycocypins inhibit three protease families.
J.Biol.Chem., 285, 2010
|
|
5X2Y
 
 | |
8QIM
 
 | | CrPhotLOV1 light state structure 42.5 ms (40-45 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
1SGC
 
 | |
8QIS
 
 | | CrPhotLOV1 light state structure 72.5 ms (70-75 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
6H6F
 
 | | PTC3 holotoxin complex from Photorhabdus luminiscens - Mutant TcC-D651A | | Descriptor: | TcdA1, TcdB2,TccC3,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
4ABN
 
 | | Crystal structure of full length mouse Strap (TTC5) | | Descriptor: | 1,2-ETHANEDIOL, TETRATRICOPEPTIDE REPEAT PROTEIN 5 | | Authors: | Pike, A.C.W, Bullock, A.N, Kleinekofort, W, Zimmermann, T, Burgess-Brown, N, Sharpe, T.D, Thangaratnarajah, C, Keates, T, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, La Thangue, N.B, Knapp, S. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P53 Cofactor Strap Exhibits an Unexpected Tpr Motif and Oligonucleotide-Binding (Ob)-Fold Structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6TA7
 
 | | CRYSTAL STRUCTURE OF HUMAN G3BP1-NTF2 IN COMPLEX WITH HUMAN CAPRIN1-DERIVED SOLOMON MOTIF | | Descriptor: | CHLORIDE ION, Caprin-1, Ras GTPase-activating protein-binding protein 1, ... | | Authors: | Schulte, T, Achour, A, Panas, M.D, McInerney, G.M. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Caprin-1 binding to the critical stress granule protein G3BP1 is regulated by pH
Biorxiv, 2021
|
|
7XX5
 
 | | Crystal Structure of Nucleosome-H1.3 Linker Histone Assembly (sticky-169a DNA fragment) | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H1.3, ... | | Authors: | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | Deposit date: | 2022-05-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment)
To Be Published
|
|
8QIG
 
 | | CrPhotLOV1 light state structure 17.5 ms (15-20 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
6HQS
 
 | | Crystal structure of GcoA F169S bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1N2Z
 
 | | 2.0 Angstrom structure of BtuF, the vitamin B12 binding protein of E. coli | | Descriptor: | CADMIUM ION, CHLORIDE ION, CYANOCOBALAMIN, ... | | Authors: | Borths, E.L, Locher, K.P, Lee, A.T, Rees, D.C. | | Deposit date: | 2002-10-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
9C1X
 
 | | Apo DUF4297 12-mer | | Descriptor: | DUF4297 domain-containing protein | | Authors: | Rish, A.D, Fosuah, E, Fu, T.M. | | Deposit date: | 2024-05-29 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Architecture remodeling activates the HerA-DUF anti-phage defense system.
Mol.Cell, 85, 2025
|
|
4QL3
 
 | | Crystal Structure of a GDP-bound G12R Oncogenic Mutant of Human GTPase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Manandhar, A, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2014-06-10 | | Release date: | 2015-06-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.041 Å) | | Cite: | Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations.
Mol Cancer Res., 13, 2015
|
|
8QIF
 
 | | CrPhotLOV1 light state structure 12.5 ms (10-15 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
1SS2
 
 | | Solution structure of the second complement control protein (CCP) module of the GABA(B)R1a receptor, Pro-119 cis conformer | | Descriptor: | Gamma-aminobutyric acid type B receptor, subunit 1 | | Authors: | Blein, S, Uhrin, D, Smith, B.O, White, J.H, Barlow, P.N. | | Deposit date: | 2004-03-23 | | Release date: | 2004-10-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the complement control protein (CCP) modules of GABA(B) receptor 1a: only one of the two CCP modules is compactly folded.
J.Biol.Chem., 279, 2004
|
|
9EWY
 
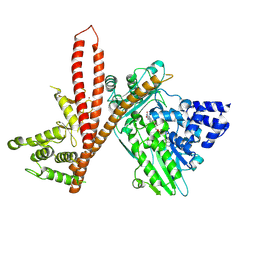 | | CryoEM structure of human MICAL1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, ZINC ION, [F-actin]-monooxygenase MICAL1 | | Authors: | Schrofel, A, Pinkas, D, Novacek, J, Rozbesky, D. | | Deposit date: | 2024-04-05 | | Release date: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of MICAL autoinhibition.
Nat Commun, 15, 2024
|
|
4QLI
 
 | |
8QIV
 
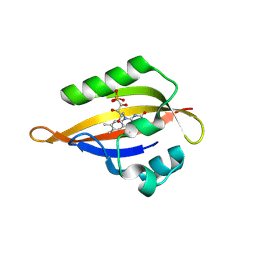 | | CrPhotLOV1 light state structure 87.5 ms (85-90 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
4EDR
 
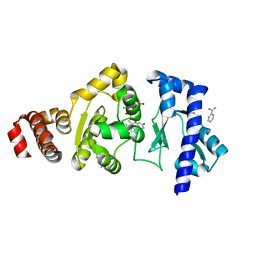 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to UTP and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, MANGANESE (II) ION, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
2RAR
 
 | |
9E71
 
 | |
6DL1
 
 | | Racemic structure of jatrophidin, an orbitide from Jatropha curcas | | Descriptor: | jatrophidin | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.029 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
