8Z9F
 
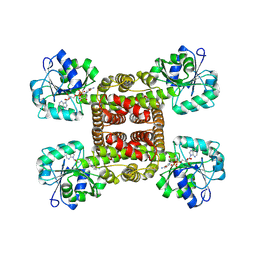 | | Crystal structure of glyoxylate reductase from Acetobacter aceti in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-hydroxyisobutyrate dehydrogenase | | Authors: | Majumder, T.R, Yoshizawa, T, Inoue, M, Aono, R, Matsumura, H, Mihara, H. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the mechanism underlying the dual cofactor specificity of glyoxylate reductase from Acetobacter aceti in the beta-hydroxyacid dehydrogenase family.
Biochim Biophys Acta Proteins Proteom, 1873, 2025
|
|
6WHU
 
 | | GluN1b-GluN2B NMDA receptor in complex with SDZ 220-040 and L689,560, class 1 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
8SVB
 
 | | Antimicrobial lasso peptide achromonodin-1 | | Descriptor: | Achromonodin-1 | | Authors: | Carson, D.V, Cheung-Lee, W.L, So, L, Link, A.J. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Discovery, Characterization, and Bioactivity of the Achromonodins: Lasso Peptides Encoded by Achromobacter .
J.Nat.Prod., 86, 2023
|
|
6UAI
 
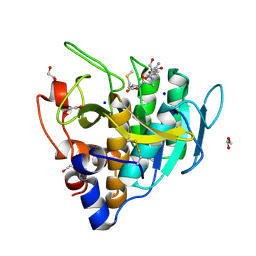 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with YSAM peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6Z82
 
 | |
6Q41
 
 | | Atomic resolution crystal structure of a BAA collagen heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Leading chain of the BAA collagen heterotrimer, ... | | Authors: | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
1NC2
 
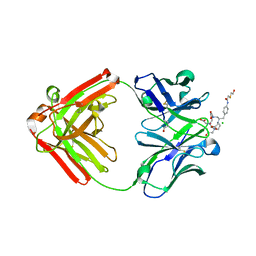 | | Crystal Structure of Monoclonal Antibody 2D12.5 Fab Complexed with Y-DOTA | | Descriptor: | (S)-2-(4-(2-(2-HYDROXYETHYLTHIO)-ACETAMIDO)-BENZYL)-1,4,7,10-TETRAAZACYCLODODECANE-N,N',N'',N'''-TETRAACETATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Corneillie, T.M, Fisher, A.J, Meares, C.F. | | Deposit date: | 2002-12-04 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two complexes of the rare-earth-DOTA-binding antibody 2D12.5: ligand generality from a chiral system.
J.Am.Chem.Soc., 125, 2003
|
|
8JUE
 
 | | Crystal structure of glutaminase C in complex with compound 11 | | Descriptor: | 2-(3-phenoxyphenyl)-N-[5-[[(3R)-1-pyridazin-3-ylpyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Wang, X, Hanyu, S, Tingting, D. | | Deposit date: | 2023-06-26 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Targeting the Subpocket Enables the Discovery of Thiadiazole-Pyridazine Derivatives as Glutaminase C Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
7QHH
 
 | | Desensitized state of GluA1/2 AMPA receptor in complex with TARP-gamma 8 (TMD-LBD) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, GLUTAMIC ACID, ... | | Authors: | Herguedas, B, Kohegyi, B, Dohrke, J.N, Watson, J.F, Zhang, D, Ho, H, Shaikh, S, Lape, R, Krieger, J.M, Greger, I.H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
6ZGL
 
 | |
8JUB
 
 | | Crystal structure of glutaminase C in complex with compound 27 | | Descriptor: | 3-[2-oxidanylidene-2-[[5-[[(3R)-1-pyridazin-3-ylpyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]amino]ethyl]benzoic acid, Glutaminase kidney isoform, mitochondrial | | Authors: | Wang, X, Hanyu, S, Tingting, D. | | Deposit date: | 2023-06-26 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Targeting the Subpocket Enables the Discovery of Thiadiazole-Pyridazine Derivatives as Glutaminase C Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8XBN
 
 | |
8K3Y
 
 | | The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Zhang, K, Chang, C.I. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
8UAI
 
 | |
6WYV
 
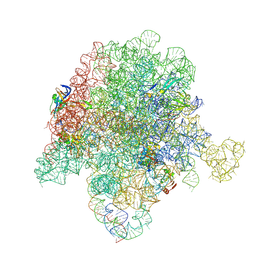 | | E. coli 50S ribosome bound to compounds 47 and VS1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
8UGB
 
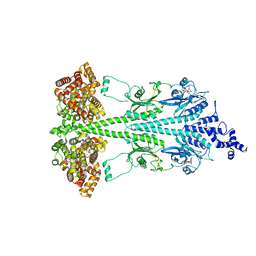 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to udenafil | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-05 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8FL8
 
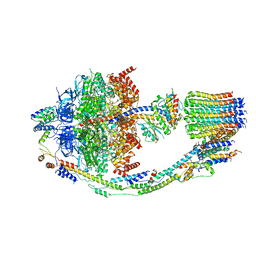 | | Yeast ATP Synthase structure in presence of MgATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7T5H
 
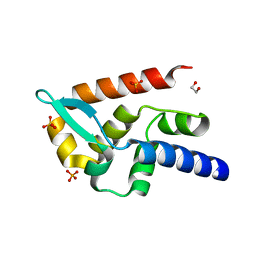 | | Structure of rabies virus phosphoprotein C-terminal domain, wild type | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phosphoprotein, ... | | Authors: | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
9AZU
 
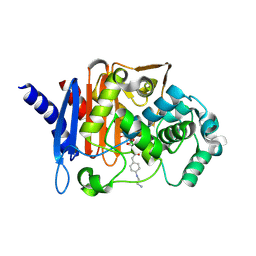 | | Crystal structure of PDC-3 beta-lactamase in complex with taniborbactam | | Descriptor: | (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Beta-lactamase | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2024-03-11 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure and mechanism of taniborbactam inhibition of the cefepime-hydrolyzing, partial R2-loop deletion Pseudomonas -derived cephalosporinase variant PDC-88.
Antimicrob.Agents Chemother., 69, 2025
|
|
8CTJ
 
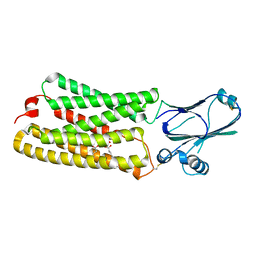 | | Cryo-EM structure of TMEM87A | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Transmembrane protein 87A | | Authors: | Hoel, C.M, Zhang, L, Brohawn, S.G. | | Deposit date: | 2022-05-15 | | Release date: | 2022-07-20 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4.74 Å) | | Cite: | Structure of the GOLD-domain seven-transmembrane helix protein family member TMEM87A.
Elife, 11, 2022
|
|
8K65
 
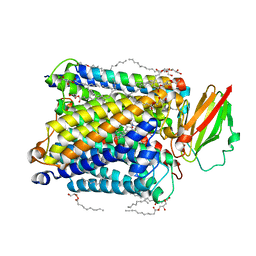 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase without pump laser irradiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Zoric, D, Sandelin, E, Nango, E, Tanaka, R, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
6O1F
 
 | |
8K6Y
 
 | | Serial femtosecond crystallography structure of photo dissociated CO from ba3- type cytochrome c oxidase determined by extrapolation method | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Zoric, D, Sandelin, E, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
8XJ2
 
 | | MAT2A in complex with SAM and allosteric inhibitor SCR-7952 | | Descriptor: | 7-chloranyl-5-(2-cyclopropylpyridin-3-yl)-1-methyl-imidazo[4,5-c]quinolin-4-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Zhou, F. | | Deposit date: | 2023-12-20 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | MAT2A in complex with SAM and allosteric inhibitor SCR-7952
To Be Published
|
|
6Y1B
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant K32A_Q126K | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
