7WJ4
 
 | |
7WIZ
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, GLUTAMINE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liu, J.L, Guo, C.J. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5O94
 
 | | X-ray structure of a zinc binding GB1 mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Rothlisberger, U, Bozkurt, E, Hovius, R, Perez, M.A.S, Browning, N.J. | | Deposit date: | 2017-06-15 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Genetic Algorithm Based Design and Experimental Characterization of a Highly Thermostable Metalloprotein.
J. Am. Chem. Soc., 140, 2018
|
|
5NJI
 
 | | Structure of the dehydratase domain of PpsC from Mycobacterium tuberculosis in complex with C12:1-CoA | | Descriptor: | Phthiocerol/phenolphthiocerol synthesis polyketide synthase type I PpsC, ~{S}-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (~{E})-dodec-2-enethioate | | Authors: | Gavalda, S, Faille, A, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into Substrate Modification by Dehydratases from Type I Polyketide Synthases.
J. Mol. Biol., 429, 2017
|
|
5NJM
 
 | | Lysozyme room-temperature structure determined by serial millisecond crystallography | | Descriptor: | Lysozyme C | | Authors: | Weinert, T, Vera, L, Marsh, M, James, D, Gashi, D, Nogly, P, Jaeger, K, Standfuss, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NLX
 
 | | A2A Adenosine receptor room-temperature structure determined by serial millisecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
8A56
 
 | | Coenzyme A-persulfide reductase (CoAPR) from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, Coenzyme A-persulfide reductase, ... | | Authors: | Costa, S.S, Walsh, B.J, Giedroc, D.P, Brito, J.A. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Metabolic and Structural Insights into Hydrogen Sulfide Mis-Regulation in Enterococcus faecalis.
Antioxidants, 11, 2022
|
|
7ZP7
 
 | | Crystal structure of evolved photoenzyme EnT1.3 (truncated) with bound product | | Descriptor: | (1~{R},10~{R},12~{S})-15-oxa-8-azatetracyclo[8.5.0.0^{1,12}.0^{2,7}]pentadeca-2(7),3,5-trien-9-one, 1,2-ETHANEDIOL, EnT1.3 C | | Authors: | Hardy, F.J, Levy, C. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A designed photoenzyme for enantioselective [2+2] cycloadditions.
Nature, 611, 2022
|
|
7ZP6
 
 | |
7ZP5
 
 | |
1AJ2
 
 | | CRYSTAL STRUCTURE OF A BINARY COMPLEX OF E. COLI DIHYDROPTEROATE SYNTHASE | | Descriptor: | DIHYDROPTEROATE SYNTHASE, SULFATE ION, [7,8-DIHYDRO-PTERIN-6-YL METHANYL]-PHOSPHONOPHOSPHATE | | Authors: | Achari, A, Somers, D.O, Champness, J.N, Bryant, P.K, Rosemond, J, Stammers, D.K. | | Deposit date: | 1997-05-14 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase.
Nat.Struct.Biol., 4, 1997
|
|
5LB9
 
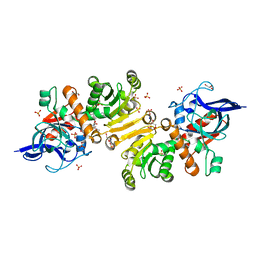 | | Structure of the T175V Etr1p mutant in the monoclinic form P21 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial, ... | | Authors: | Wagner, T, Rosenthal, R.G, Voegeli, B, Shima, S, Erb, T.J. | | Deposit date: | 2016-06-15 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A conserved threonine prevents self-intoxication of enoyl-thioester reductases.
Nat. Chem. Biol., 13, 2017
|
|
5L53
 
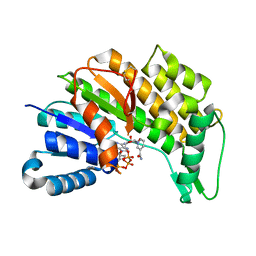 | | Menthone neomenthol reductase from Mentha piperita in complex with NADP | | Descriptor: | (-)-menthone:(+)-neomenthol reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5KZ5
 
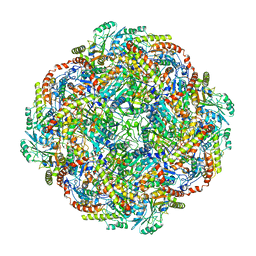 | | Architecture of the Human Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Complex Formed by the Iron Donor, the Sulfur Donor, and the Scaffold | | Descriptor: | Cysteine desulfurase, mitochondrial, Frataxin, ... | | Authors: | Gakh, O, Ranatunga, W, Smith, D.Y, Ahlgren, E.C, Al-Karadaghi, S, Thompson, J.R, Isaya, G. | | Deposit date: | 2016-07-22 | | Release date: | 2016-08-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (14.3 Å) | | Cite: | Architecture of the Human Mitochondrial Iron-Sulfur Cluster Assembly Machinery.
J.Biol.Chem., 291, 2016
|
|
5L84
 
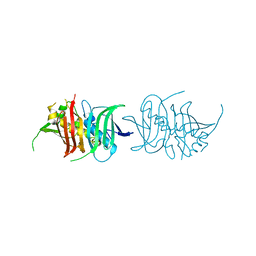 | |
5L3B
 
 | | Human LSD1/CoREST: LSD1 D556G mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
5L3D
 
 | | Human LSD1/CoREST: LSD1 Y761H mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
1B66
 
 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | Descriptor: | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE, BIOPTERIN, ZINC ION | | Authors: | Ploom, T, Thoeny, B, Yim, J, Lee, S, Nar, H, Leimbacher, W, Huber, R, Richardson, J, Auerbach, G. | | Deposit date: | 1999-01-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and kinetic investigations on the mechanism of 6-pyruvoyl tetrahydropterin synthase.
J.Mol.Biol., 286, 1999
|
|
5LBX
 
 | | Structure of the T175V Etr1p mutant in the trigonal form P312 in complex with NADP and crotonyl-CoA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Enoyl-[acyl-carrier-protein] reductase [NADPH, ... | | Authors: | Wagner, T, Rosenthal, R.G, Voegeli, B, Shima, S, Erb, T.J. | | Deposit date: | 2016-06-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved threonine prevents self-intoxication of enoyl-thioester reductases.
Nat. Chem. Biol., 13, 2017
|
|
5L4S
 
 | | Isopiperitenone reductase from Mentha piperita in complex with NADP and beta-Cyclocitral | | Descriptor: | (-)-isopiperitenone reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, beta-cyclocitral | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-05-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5L51
 
 | |
1C3E
 
 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLATE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 2-{4-[2-(2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL)-1-CARBOXY-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-27 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
5L3C
 
 | | Human LSD1/CoREST: LSD1 E379K mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
1BRM
 
 | | ASPARTATE BETA-SEMIALDEHYDE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE | | Authors: | Hadfield, A.T, Kryger, G, Ouyang, J, Ringe, D, Petsko, G.A, Viola, R.E. | | Deposit date: | 1998-08-24 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of aspartate-beta-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis.
J.Mol.Biol., 289, 1999
|
|
1CI7
 
 | | TERNARY COMPLEX OF THYMIDYLATE SYNTHASE FROM PNEUMOCYSTIS CARINII | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Anderson, A.C, O'Neil, R.H, Delano, W.L, Stroud, R.M. | | Deposit date: | 1999-04-08 | | Release date: | 2000-04-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural mechanism for half-the-sites reactivity in an enzyme, thymidylate synthase, involves a relay of changes between subunits.
Biochemistry, 38, 1999
|
|
