3T96
 
 | | Iodowillardiine bound to a double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Wang, S, Chuang, H.H, Oswald, R.E. | | Deposit date: | 2011-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Mechanism of AMPA Receptor Activation by Partial Agonists: DISULFIDE TRAPPING OF CLOSED LOBE CONFORMATIONS.
J.Biol.Chem., 286, 2011
|
|
3T9H
 
 | | Kainate bound to a double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Wang, S, Chuang, H.H, Oswald, R.E. | | Deposit date: | 2011-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Mechanism of AMPA Receptor Activation by Partial Agonists: DISULFIDE TRAPPING OF CLOSED LOBE CONFORMATIONS.
J.Biol.Chem., 286, 2011
|
|
2AVT
 
 | | Crystal structure of the beta subunit from DNA polymerase of Streptococcus pyogenes | | Descriptor: | DNA polymerase III beta subunit | | Authors: | Argiriadi, M.A, Goedken, E.R, Bruck, I, O'donnell, M, Kuriyan, J. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium.
Bmc Struct.Biol., 6, 2006
|
|
3UA8
 
 | | Crystal Structure Analysis of a 6-Amino Quinazolinedione Sulfonamide bound to human GluR2 | | Descriptor: | Glutamate receptor 2, N-methyl-1-{3-[(methylsulfonyl)amino]-2,4-dioxo-7-(trifluoromethyl)-1,2,3,4-tetrahydroquinazolin-6-yl}-1H-imidazole-4-carboxamide | | Authors: | Kallen, J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 6-Amino quinazolinedione sulfonamides as orally active competitive AMPA receptor antagonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3TM0
 
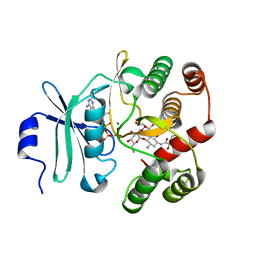 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa AMPPNP Butirosin A Complex | | Descriptor: | (2S)-4-amino-N-[(1R,2S,3R,4R,5S)-5-amino-4-[(2,6-diamino-2,6-dideoxy-alpha-D-glucopyranosyl)oxy]-2-hydroxy-3-(beta-D-xylofuranosyloxy)cyclohexyl]-2-hydroxybutanamide, Aminoglycoside 3'-phosphotransferase, MAGNESIUM ION, ... | | Authors: | Berghuis, A.M, Fong, D.H. | | Deposit date: | 2011-08-30 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of APH(3')-IIIa-mediated resistance to N1-substituted aminoglycoside antibiotics.
Antimicrob.Agents Chemother., 53, 2009
|
|
7PRD
 
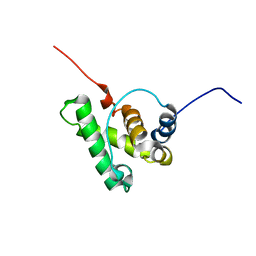 | |
7PRE
 
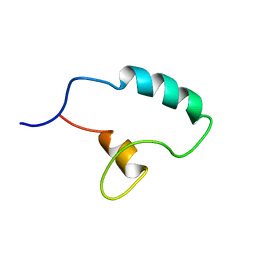 | |
7QJI
 
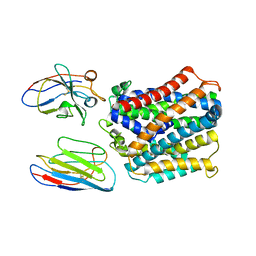 | | X-Ray Structure of apo-EleNRMT in complex with two Nanobodies at 4.1A | | Descriptor: | Divalent metal cation transporter, Elen-Nanobody-complex | | Authors: | Ramanadane, K, Straub, M.S, Dutzler, R, Manatschal, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural and functional properties of a magnesium transporter of the SLC11/NRAMP family.
Elife, 11, 2022
|
|
7QJJ
 
 | | X-Ray Structure of a Mn2+ soak of EleNRMT in complex with two Nanobodies at 4.6A | | Descriptor: | Divalent metal cation transporter, Elen-Nb1-Nb2, MANGANESE (II) ION | | Authors: | Ramanadane, K, Straub, M.S, Dutzler, R, Manatschal, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural and functional properties of a magnesium transporter of the SLC11/NRAMP family.
Elife, 11, 2022
|
|
3KZO
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate and N-acetyl-L-norvaline | | Descriptor: | GLYCEROL, N-ACETYL-L-NORVALINE, N-acetylornithine carbamoyltransferase, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
3KZM
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate | | Descriptor: | GLYCEROL, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
3SM2
 
 | | The crystal structure of XMRV protease complexed with Amprenavir | | Descriptor: | gag-pro-pol polyprotein, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.
Febs J., 278, 2011
|
|
3KZN
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with N-acetyl-L-ornirthine | | Descriptor: | GLYCEROL, N-acetylornithine carbamoyltransferase, N~2~-ACETYL-L-ORNITHINE, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
2AO7
 
 | | Adam10 Disintegrin and cysteine- rich domain | | Descriptor: | ADAM 10, SULFATE ION | | Authors: | Janes, P.W, Saha, N, Barton, W.A, Kolev, M.V, Wimmer-Kleikamp, S.H, Nievergall, E, Blobel, C.P, Himanen, J.-P, Lackmann, M, Nikolov, D.B. | | Deposit date: | 2005-08-12 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans.
Cell(Cambridge,Mass.), 123, 2005
|
|
8SQ9
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8SQJ
 
 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8SQK
 
 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8FYN
 
 | | MicroED structure of A2A from plasma milled lamellae | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Martynowycz, M.W, Shiriaeva, A, Clabbers, M.T.B, Nicolas, W.J, Weaver, S.J, Hattne, J, Gonen, T. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals.
Nat Commun, 14, 2023
|
|
4P3X
 
 | | Structure of the Fe4S4 quinolinate synthase NadA from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Cherrier, M.V, Chan, A, Darnault, C, Reichmann, D, Amara, P, Ollagnier de Choudens, S, Fontecilla-Camps, J.C. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of Fe4S4 quinolinate synthase unravels an enzymatic dehydration mechanism that uses tyrosine and a hydrolase-type triad.
J.Am.Chem.Soc., 136, 2014
|
|
2FQ5
 
 | |
7QNG
 
 | | Structure of a MHC I-Tapasin-ERp57 complex | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Mueller, I.K, Thomas, C, Trowitzsch, S, Tampe, R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an MHC I-tapasin-ERp57 editing complex defines chaperone promiscuity.
Nat Commun, 13, 2022
|
|
2FH9
 
 | |
5EGM
 
 | | Development of a novel tricyclic class of potent and selective FIXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-chloranyl-~{N}-[(7~{S})-2-methyl-7-phenyl-10-(1~{H}-1,2,3,4-tetrazol-5-yl)-8,9-dihydro-6~{H}-pyrido[1,2-a]indol-7-yl]-4-(1,2,4-triazol-4-yl)benzamide, Coagulation factor IX, ... | | Authors: | Meng, D, Andre, P, Bateman, T.J, Berger, R, Chen, Y, Desai, K, Dewnani, S, Ellsworth, K, Feng, D, Geissler, W.M, Guo, L, Hruza, A, Jian, T, Li, H, Parker, D.L, Reichert, P, Sherer, E.C, Smith, C.J, Sonatore, L.M, Tschirret-Guth, R, Wu, J, Xu, J, Zhang, T, Campeau, L, Orr, R, Poirier, M, McCabe-Dunn, j, Araki, K, Nishimura, T, Sakurada, I, Hirabayashi, T, Wood, H.B. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Development of a novel tricyclic class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5E2G
 
 | | Crystal Structure of D-alanine Carboxypeptidase AmpC from Burkholderia cenocepacia | | Descriptor: | ACETIC ACID, Beta-lactamase, THIOCYANATE ION | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal Structure of D-alanine Carboxypeptidase AmpC from Burkholderia cenocepacia
To Be Published
|
|
1FSY
 
 | | AMPC BETA-LACTAMASE FROM E. COLI COMPLEXED WITH INHIBITOR CLOXACILLINBORONIC ACID | | Descriptor: | CEPHALOSPORINASE, N-[5-METHYL-3-O-TOLYL-ISOXAZOLE-4-CARBOXYLIC ACID AMIDE] BORONIC ACID, PHOSPHATE ION | | Authors: | Caselli, E, Powers, R.A, Blasczcak, L.C, Wu, C.Y, Prati, F, Shoichet, B.K. | | Deposit date: | 2000-09-11 | | Release date: | 2001-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Energetic, structural, and antimicrobial analyses of beta-lactam side chain recognition by beta-lactamases.
Chem.Biol., 8, 2001
|
|
