7K3L
 
 | |
4WRQ
 
 | |
1Q9X
 
 | | Crystal structure of Enterobacteria phage RB69 gp43 DNA polymerase complexed with tetrahydrofuran containing DNA | | Descriptor: | 1',2'-DIDEOXYRIBOFURANOSE-5'-PHOSPHATE, 2',3'-DIDEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Freisinger, E, Grollman, A.P, Miller, H, Kisker, C. | | Deposit date: | 2003-08-26 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Lesion (in)tolerance reveals insights into DNA replication fidelity.
Embo J., 23, 2004
|
|
4XW6
 
 | | X-ray structure of PKAc with ADP, free phosphate ion, CP20, magnesium ions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
4XW5
 
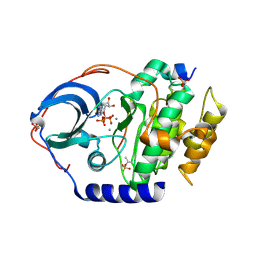 | | X-ray structure of PKAc with ATP, CP20, calcium ions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
6DMG
 
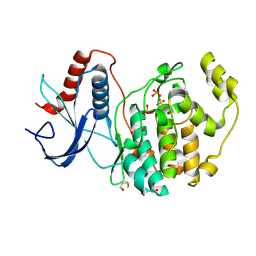 | | A multiconformer ligand model of EK6 bound to ERK2 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Hudson, B.M, van Zundert, G.C.P, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
5BQ8
 
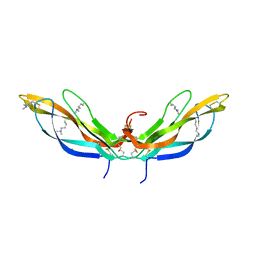 | | Crystal structure of Norrin, a Wnt signalling activator, Crystal Form II | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Norrin | | Authors: | Chang, T.-H, Hsieh, F.-L, Harlos, K, Jones, E.Y. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan.
Elife, 4, 2015
|
|
4XW4
 
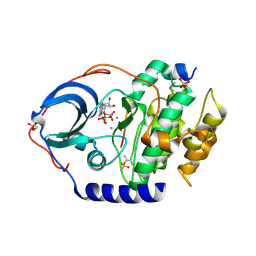 | | X-ray structure of PKAc with AMPPNP, SP20, calcium ions | | Descriptor: | CALCIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
6VCD
 
 | | Cryo-EM structure of IRP2-FBXL5-SKP1 complex | | Descriptor: | F-box/LRR-repeat protein 5, FE2/S2 (INORGANIC) CLUSTER, Iron-responsive element binding protein 2, ... | | Authors: | Wang, H, Shi, H, Zheng, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2Fe2S] Cluster.
Mol.Cell, 78, 2020
|
|
5VPX
 
 | | I38T mutant of 2009 H1N1 PA Endonuclease in complex with RO-7 | | Descriptor: | 1-[(11S)-6,11-dihydrodibenzo[b,e]thiepin-11-yl]-5-hydroxy-3-[(2R)-1,1,1-trifluoropropan-2-yl]-2,3-dihydro-1H-pyrido[2,1-f][1,2,4]triazine-4,6-dione, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-06 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
5VQN
 
 | | E119D mutant of 2009 H1N1 PA Endonuclease in complex with RO-7 | | Descriptor: | 1-[(11S)-6,11-dihydrodibenzo[b,e]thiepin-11-yl]-5-hydroxy-3-[(2R)-1,1,1-trifluoropropan-2-yl]-2,3-dihydro-1H-pyrido[2,1-f][1,2,4]triazine-4,6-dione, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-09 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
1FRP
 
 | | CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE-2,6-BISPHOSPHATE, AMP AND ZN2+ AT 2.0 ANGSTROMS RESOLUTION. ASPECTS OF SYNERGISM BETWEEN INHIBITORS | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Xue, Y, Huang, S, Liang, J.-Y, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-08-26 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 2,6-bisphosphate, AMP, and Zn2+ at 2.0-A resolution: aspects of synergism between inhibitors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1FPB
 
 | | CRYSTAL STRUCTURE OF THE NEUTRAL FORM OF FRUCTOSE 1,6-BISPHOSPHATASE COMPLEXED WITH REGULATORY INHIBITOR FRUCTOSE 2,6-BISPHOSPHATE AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Liang, J.Y, Huang, S, Zhang, Y, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the neutral form of fructose 1,6-bisphosphatase complexed with regulatory inhibitor fructose 2,6-bisphosphate at 2.6-A resolution.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
5VPT
 
 | | 2009 H1N1 PA Endonuclease in complex with RO-7 | | Descriptor: | 1-[(11S)-6,11-dihydrodibenzo[b,e]thiepin-11-yl]-5-hydroxy-3-[(2R)-1,1,1-trifluoropropan-2-yl]-2,3-dihydro-1H-pyrido[2,1-f][1,2,4]triazine-4,6-dione, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-05 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
5VP8
 
 | | I38T mutant of 2009 H1N1 PA Endonuclease | | Descriptor: | 1-[(3R,5S,7R)-1,5,7,9-tetrakis(2-oxopyrrolidin-1-yl)nonan-3-yl]-1,3-dihydro-2H-pyrrol-2-one, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-04 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
8S0F
 
 | | H. sapiens OC1M bound to double stranded DNA | | Descriptor: | DNA (39-mer), DNA replication factor Cdt1, DNA replication licensing factor MCM2, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | MCM double hexamer loading visualized with human proteins.
Nature, 636, 2024
|
|
8S0E
 
 | | H. sapiens OCCM bound to double stranded DNA | | Descriptor: | Cell division control protein 6 homolog, DNA (39-mer), DNA replication factor Cdt1, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | MCM double hexamer loading visualized with human proteins.
Nature, 636, 2024
|
|
6M93
 
 | | Monophosphorylated pSer33 b-Catenin peptide, b-TrCP/Skp1, NRX-1933 ternary complex | | Descriptor: | 2-oxo-N-[3-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)-1,2-dihydropyridine-3-carboxamide, Catenin beta-1, F-box/WD repeat-containing protein 1A, ... | | Authors: | Simonetta, K.R, Clifton, M.C, Walter, R.L, Ranieri, G.M, Lee, S.J. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction.
Nat Commun, 10, 2019
|
|
8TW9
 
 | | Cryo-EM structure of S. cerevisiae PolE-Ctf18-8-1-DNA | | Descriptor: | Chromosome transmission fidelity protein 18, Chromosome transmission fidelity protein 8, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis.
Science, 385, 2024
|
|
6M92
 
 | | Monophosphorylated pSer33 b-Catenin peptide, b-TrCP/Skp1, NRX-2663 ternary complex | | Descriptor: | 3-{[2-oxo-4-phenoxy-6-(trifluoromethyl)-1,2-dihydropyridine-3-carbonyl]amino}benzoic acid, Catenin beta-1, F-box/WD repeat-containing protein 1A, ... | | Authors: | Simonetta, K.R, Clifton, M.C, Walter, R.L, Ranieri, G.M, Carter, J.J, Lee, S.J. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction.
Nat Commun, 10, 2019
|
|
3OXL
 
 | | Human lysine methyltransferase Smyd3 in complex with AdoHcy (Form II) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 3, ZINC ION | | Authors: | Xu, S, Wu, J, Sun, B, Zhong, C, Ding, J. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding
Nucleic Acids Res., 39, 2011
|
|
4NAC
 
 | |
7UQI
 
 | |
7UQK
 
 | |
3O96
 
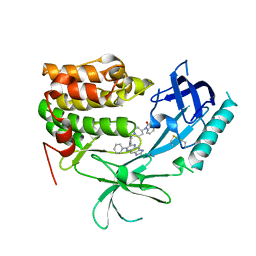 | | Crystal Structure of Human AKT1 with an Allosteric Inhibitor | | Descriptor: | 1-(1-(4-(7-phenyl-1H-imidazo[4,5-g]quinoxalin-6-yl)benzyl)piperidin-4-yl)-1H-benzo[d]imidazol-2(3H)-one, RAC-alpha serine/threonine-protein kinase | | Authors: | Voegtli, W.C, Wu, W.-I, Lord-Ondash, H.A, Dizon, F.P, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition.
Plos One, 5, 2010
|
|
