8EJP
 
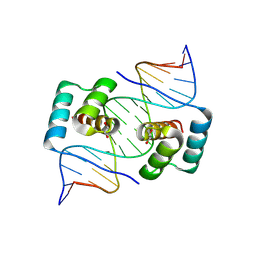 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA containing 5-Bromouracil | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Yin, L, Shi, K, Aihara, H. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
7O82
 
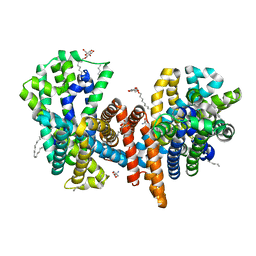 | | The L-arginine/agmatine antiporter from E. coli at 1.7 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine/agmatine antiporter, DECANE, ... | | Authors: | Jeckelmann, J.M, Ilgue, H, Kalbermatter, D, Fotiadis, D. | | Deposit date: | 2021-04-14 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution structure of the amino acid transporter AdiC reveals insights into the role of water molecules and networks in oligomerization and substrate binding.
Bmc Biol., 19, 2021
|
|
8EJO
 
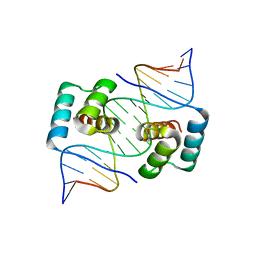 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Yin, L, Shi, K, Aihara, H. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
7OJY
 
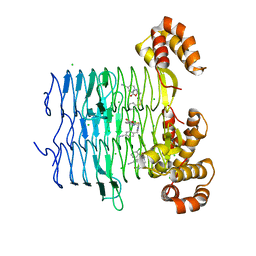 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 6 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-[(5-oxidanyl-1,3,4-oxadiazol-2-yl)methyl]ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OMN
 
 | | Anti-EphA1 JD1-1 VH domain | | Descriptor: | GLYCEROL, IMIDAZOLE, JD1-1 VH domain | | Authors: | Ereno Orbea, J, Nilvebrant, J, Sidhu, S, Julien, J.P. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Systematic Engineering of Optimized Autonomous Heavy-Chain Variable Domains.
J.Mol.Biol., 433, 2021
|
|
7OK1
 
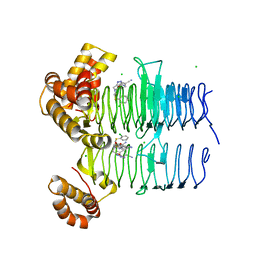 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 3 | | Descriptor: | Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, ~{N}-[(5-azanyl-1,3,4-oxadiazol-2-yl)methyl]-2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]ethanamide | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
6VRB
 
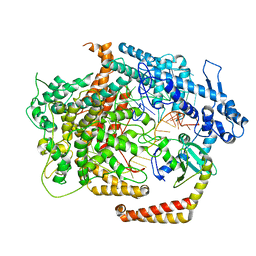 | | Cryo-EM structure of AcrVIA1-Cas13(crRNA) complex | | Descriptor: | AcrVIA1, CRISPR-associated endoribonuclease Cas13a, RNA (52-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
6MXF
 
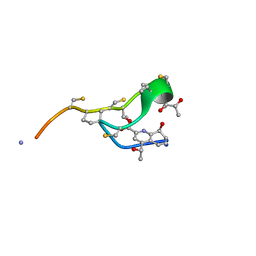 | | MicroED structure of thiostrepton at 1.9 A resolution | | Descriptor: | Thiostrepton | | Authors: | Jones, C.G, Martynowycz, M.W, Hattne, J, Fulton, T, Stoltz, B.M, Rodriguez, J.A, Nelson, H.M, Gonen, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.91 Å) | | Cite: | The CryoEM Method MicroED as a Powerful Tool for Small Molecule Structure Determination.
ACS Cent Sci, 4, 2018
|
|
3CAY
 
 | | Crystal structure of Lipopeptide Detergent (LPD-12) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, LPD-12 | | Authors: | Ho, D.N, Pomroy, N.C, Cuesta-Seijo, J.A, Prive, G.G. | | Deposit date: | 2008-02-20 | | Release date: | 2008-09-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a self-assembling lipopeptide detergent at 1.20 A.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5KR4
 
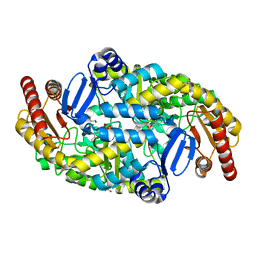 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
1EPR
 
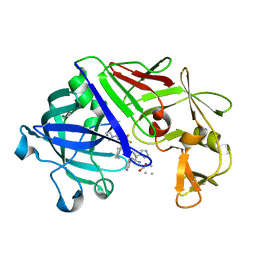 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-135,040 | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Badasso, M, Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
8YGV
 
 | | F1 domain of Non-catalytic site depleted and epsilon C-terminal domain deleted FoF1-ATPase from Bacillus PS3,nucleotide depleted condition | | Descriptor: | ATP synthase gamma chain, ATP synthase subunit alpha, ATP synthase subunit beta | | Authors: | Kobayashi, R, Nakano, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2024-02-27 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | ADP-inhibited structure of non-catalytic site-depleted F o F 1 -ATPase from thermophilic Bacillus sp. PS-3.
Biochim Biophys Acta Bioenerg, 1866, 2025
|
|
6VRC
 
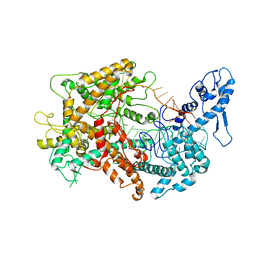 | | Cryo-EM structure of Cas13(crRNA) | | Descriptor: | CRISPR-associated endoribonuclease Cas13a, RNA (51-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
7O04
 
 | |
9FNU
 
 | |
6W98
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, CALCIUM ION, ... | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-22 | | Release date: | 2020-05-13 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
8RD3
 
 | |
8YH8
 
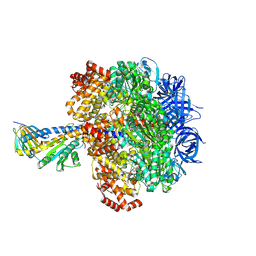 | | F1 domain of Non-catalytic site depleted and epsilon C-terminal domain deleted FoF1-ATPase from Bacillus PS3,under ATP saturated condition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Kobayashi, R, Nakano, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2024-02-27 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | ADP-inhibited structure of non-catalytic site-depleted F o F 1 -ATPase from thermophilic Bacillus sp. PS-3.
Biochim Biophys Acta Bioenerg, 1866, 2025
|
|
7O9Y
 
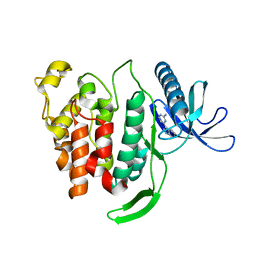 | |
7OPB
 
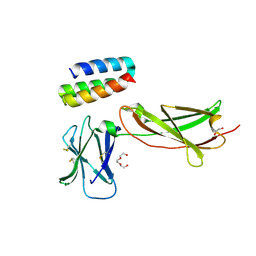 | | IL7R in complex with an antagonist | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IL7R binder, ... | | Authors: | Markovic, I, Verschueren, K.H.G, Verstraete, K, Savvides, S.N. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
6VIU
 
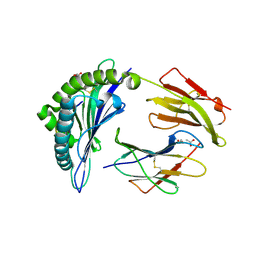 | |
8FDR
 
 | | Structure of a dodecamer complex: 5'-CGCGAAAAGCCG-3-DB1476 | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*TP*TP*TP*CP*GP*CP*G)-3'), ... | | Authors: | Ogbonna, E, Wilson, W.D. | | Deposit date: | 2022-12-04 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
6VVC
 
 | |
8Z75
 
 | | The structure of non-activated thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-04-19 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Effect of Copper Ions on the Crystal Packing and Conformation of Thiocyanate Dehydrogenase in the Crystal Structure
Crystallography Reports, 70, 2025
|
|
7OL5
 
 | |
