7MA1
 
 | | HIV-1 Protease (I84V) in Complex with GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with GRL-98065
To Be Published
|
|
8VCB
 
 | |
5VJM
 
 | | Crystal structure of H7 hemagglutinin mutant (V186K, K193T, G228S) from the influenza virus A/Shanghai/2/2013 (H7N9) with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2017-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Three mutations switch H7N9 influenza to human-type receptor specificity.
PLoS Pathog., 13, 2017
|
|
5VQJ
 
 | |
7MA0
 
 | | HIV-1 Protease (I84V) in Complex with LR2-91 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[4-(hydroxymethyl)phenyl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with LR2-91
To Be Published
|
|
8VXL
 
 | | Crystal Structure of the External Aldimine Complex of Pyridoxal-5'-Tetrazole and (S)-4-amino-5-phenoxypentanoate with the Bacillus subtilis GabR C-terminal Effector-binding and Oligomerization Domain | | Descriptor: | (4S)-4-[(E)-({3-hydroxy-2-methyl-5-[(E)-2-(1H-tetrazol-5-yl)ethenyl]pyridin-4-yl}methylidene)amino]-5-phenoxypentanoic acid, GLYCEROL, HTH-type transcriptional regulatory protein GabR, ... | | Authors: | Kaley, N.E, Liveris, Z.J, Becker, D.P, Liu, D. | | Deposit date: | 2024-02-05 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Bioisosteric replacement of pyridoxal-5'-phosphate to pyridoxal-5'-tetrazole targeting Bacillus subtilis GabR.
Protein Sci., 34, 2025
|
|
5I6W
 
 | |
5VQR
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7MA2
 
 | | HIV-1 Protease (I84V) in Complex with a Darunavir Derivative | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(1,3-benzothiazol-6-yl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.869 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with a Darunavir Derivative
To Be Published
|
|
8VSQ
 
 | | Crystal structure of Esub1 | | Descriptor: | Esub1, SULFATE ION | | Authors: | Muna, T.H, Prehna, G. | | Deposit date: | 2024-01-24 | | Release date: | 2025-01-08 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The phage protein paratox is a multifunctional metabolic regulator of Streptococcus.
Nucleic Acids Res., 53, 2025
|
|
5I72
 
 | | Crystal structure of the oligomeric form of the Lassa virus matrix protein Z | | Descriptor: | RING finger protein Z, ZINC ION | | Authors: | Hastie, K, Zandonatti, M, Liu, T, Li, S, Woods Jr, V, Saphire, E.O. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Oligomeric Form of Lassa Virus Matrix Protein Z.
J.Virol., 90, 2016
|
|
5ID6
 
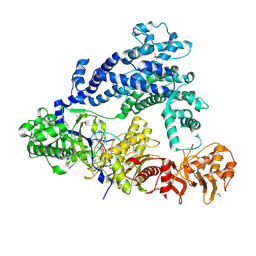 | | Structure of Cpf1/RNA Complex | | Descriptor: | Cpf1, MAGNESIUM ION, RNA (5'-R(P*AP*AP*UP*UP*UP*CP*UP*AP*CP*UP*AP*AP*GP*UP*GP*UP*AP*GP*AP*UP*C)-3') | | Authors: | Dong, D, Ren, K, Qiu, X, Wang, J, Huang, Z. | | Deposit date: | 2016-02-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The crystal structure of Cpf1 in complex with CRISPR RNA
Nature, 532, 2016
|
|
8VXK
 
 | |
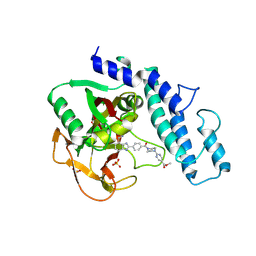
6VKO
 
 | |
5VQZ
 
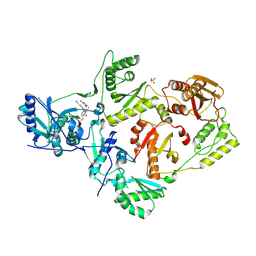 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) Variant in Complex with 2-chloro-N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacetamide (JLJ686), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Buckingham, A.B, Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7M1S
 
 | |
5IDO
 
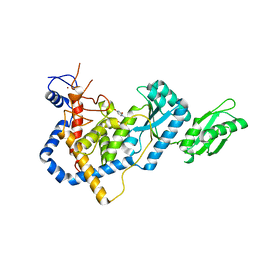 | | RNA Editing TUTase 1 from Trypanosoma brucei in complex with UTP | | Descriptor: | 3' terminal uridylyl transferase, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
7MAF
 
 | | HIV-1 Protease (I84V) in Complex with PU2 (LR2-79) | | Descriptor: | Protease, SULFATE ION, diethyl ({4-[(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}butyl]phenoxy}methyl)phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with PU2 (LR2-79)
To Be Published
|
|
7M9G
 
 | | HIV-1 Protease (I84V) in Complex with LR2-18 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with LR2-18
To Be Published
|
|
5IDT
 
 | |
6VLU
 
 | | Factor XIa in complex with compound 7 | | Descriptor: | CITRIC ACID, Coagulation factor XIa light chain, N-cyclohexyl-D-leucyl-N-[(1-aminoisoquinolin-6-yl)methyl]-4,4-difluoro-L-prolinamide | | Authors: | Lesburg, C.A, Fradera, X. | | Deposit date: | 2020-01-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Augmenting Hit Identification by Virtual Screening Techniques in Small Molecule Drug Discovery.
J.Chem.Inf.Model., 60, 2020
|
|
8VFS
 
 | |
8VG6
 
 | | Structure of Tulane virus | | Descriptor: | Capsid protein | | Authors: | Yu, G, Li, K, Jiang, W. | | Deposit date: | 2023-12-22 | | Release date: | 2024-12-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of Tulane virus
To Be Published
|
|
7M9H
 
 | | HIV-1 Protease (I84V) in Complex with LR2-20 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with LR2-20
To Be Published
|
|
5IE6
 
 | | Crystal structure of a lactonase mutant in complex with substrate b | | Descriptor: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
