7Y4U
 
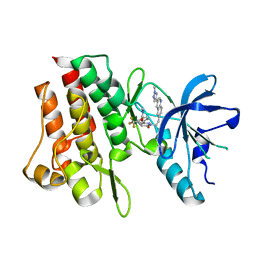 | | Crystal structure of cMET kinase domain bound by compound 9Y | | Descriptor: | Hepatocyte growth factor receptor, ~{N}-methyl-4-[1-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-oxidanylidene-pyridazin-3-yl]-2-(trifluoromethyl)benzamide | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
5VN9
 
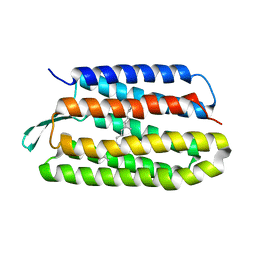 | | Structure of bacteriorhodopsin from crystals grown at 4 deg C using GlyNCOC15+4 as an LCP host lipid | | Descriptor: | Bacteriorhodopsin | | Authors: | Ishchenko, A, Peng, L, Zinovev, E, Vlasov, A, Lee, S.C, Kuklin, A, Mishin, A, Borshchevskiy, V, Zhang, Q, Cherezov, V. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Chemically Stable Lipids for Membrane Protein Crystallization.
Cryst Growth Des, 17, 2017
|
|
5VXR
 
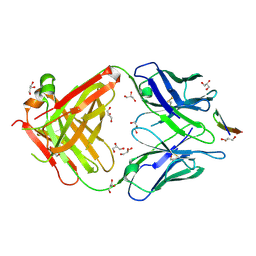 | | The antigen-binding fragment of MAb24 in complex with a peptide from Hepatitis C Virus E2 epitope I (412-423) | | Descriptor: | GLYCEROL, MAb24 Variable Heavy Chain,MAb24 Variable Heavy Chain, MAb24 Variable Light Chain,MAb24 Variable Light Chain, ... | | Authors: | Hardy, J.M, Gu, J, Boo, I, Drummer, H.E, Coulibaly, F.J. | | Deposit date: | 2017-05-24 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Escape of Hepatitis C Virus from Epitope I Neutralization Increases Sensitivity of Other Neutralization Epitopes.
J. Virol., 92, 2018
|
|
1IK5
 
 | | Crystal Structure of a 14mer RNA Containing Double UU Bulges in Two Crystal Forms: A Novel U*(AU) Intramolecular Base Triple | | Descriptor: | 5'-R(*GP*GP*UP*AP*UP*UP*UP*CP*GP*GP*UP*AP*(CBR)P*C)-3', 5'-R(*GP*GP*UP*AP*UP*UP*UP*UP*GP*GP*UP*AP*(CBR)P*C)-3', MAGNESIUM ION | | Authors: | Deng, J, Xiong, Y, Sudarsanakumar, C, Shi, K, Sundaralingam, M. | | Deposit date: | 2001-05-02 | | Release date: | 2001-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two forms of a 14-mer RNA/DNA chimer duplex with double UU bulges: a novel intramolecular U*(A x U) base triple.
RNA, 7, 2001
|
|
3TGU
 
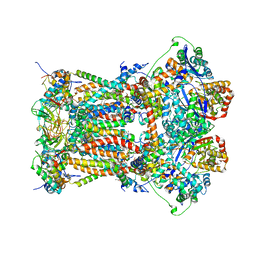 | | Cytochrome bc1 complex from chicken with pfvs-designed moa inhibitor bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | Authors: | Huang, L.-S, Yang, G.-F, Berry, E.A. | | Deposit date: | 2011-08-17 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Computational discovery of picomolar Q(o) site inhibitors of cytochrome bc1 complex.
J.Am.Chem.Soc., 134, 2012
|
|
2NUV
 
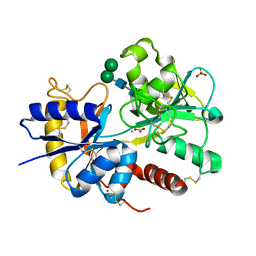 | | Crystal structure of the complex of C-terminal lobe of bovine lactoferrin with atenolol at 2.25 A resolution | | Descriptor: | 2-(4-(2-HYDROXY-3-(ISOPROPYLAMINO)PROPOXY)PHENYL)ETHANAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Singh, N, Sinha, M, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2006-11-10 | | Release date: | 2006-12-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the complex of C-terminal lobe of bovine lactoferrin with atenolol at 2.25 A resolution
To be Published
|
|
4MSW
 
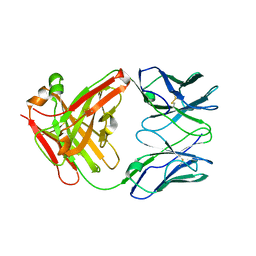 | | Y78 ester mutant of KcsA in high K+ | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, DIACYL GLYCEROL, ... | | Authors: | Matulef, K, Valiyaveetil, F.I. | | Deposit date: | 2013-09-18 | | Release date: | 2013-10-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Using protein backbone mutagenesis to dissect the link between ion occupancy and C-type inactivation in K+ channels.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2F9U
 
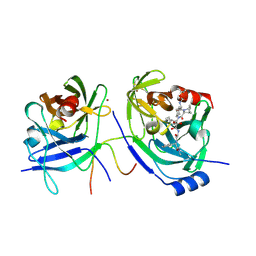 | | HCV NS3 protease domain with NS4a peptide and a ketoamide inhibitor with a P2 norborane | | Descriptor: | 1,1-DIMETHYLETHYL [1-CYCLOHEXYL-2-[3-[[[1-[2-[[2-[[2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO]-2-OXOETHYL]AMINO]-1,2-DIOXOETHYL]PENTYL]AMINO]CARBONYL]-2-AZABICYCLO[2.2.1]HEPTAN-2-YL]-2-OXOETHYL]CARBAMATE, NS3 protease/helicase', ZINC ION, ... | | Authors: | Venkatraman, S, Njoroge, F.G, Wu, W, Girijavallabhan, V, Prongay, A.J, Butkiewicz, N, Pichardo, J. | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel Inhibitors of Hepatitis C NS3-NS4A Serine Protease Derived from 2-Aza-bicyclo[2.2.1]heptane-3-carboxylic acid.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3Q6W
 
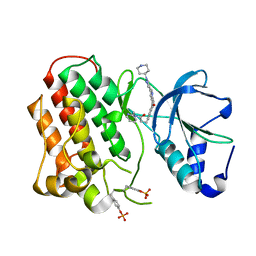 | | Structure of dually-phosphorylated MET receptor kinase in complex with an MK-2461 analog with specificity for the activated receptor | | Descriptor: | 3-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}-N-(pyridin-2-ylmethyl)propanamide, Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Rickert, K.W, Patel, S.B, Lumb, K.J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selective small molecule kinase inhibition of activated c-Met.
J.Biol.Chem., 286, 2011
|
|
7Z2I
 
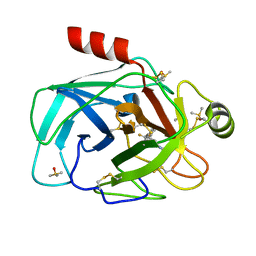 | | TRYPSIN (BOVINE) COMPLEXED WITH compound 4 | | Descriptor: | 5-[[3-(trifluoromethyl)phenyl]methyl]-1,4,6,7-tetrahydroimidazo[4,5-c]pyridine, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schiering, N, Dalvit, C, Vulpetti, A. | | Deposit date: | 2022-02-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Efficient Screening of Target-Specific Selected Compounds in Mixtures by 19 F NMR Binding Assay with Predicted 19 F NMR Chemical Shifts.
Chemmedchem, 17, 2022
|
|
1DXW
 
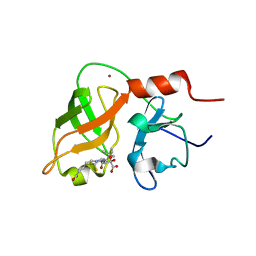 | | structure of hetero complex of non structural protein (NS) of hepatitis C virus (HCV) and synthetic peptidic compound | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Cordier, F, Narjes, F, Gerlach, B, Sambucini, S, Grzesiek, S, Matassa, V.G, Defrancesco, R, Bazzo, R. | | Deposit date: | 2000-01-17 | | Release date: | 2001-01-12 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Inhibitor Binding Induces Active Site Stabilisation of the Hcv Ns3 Protein Serine Protease Domain
Embo J., 19, 2000
|
|
3LIS
 
 | |
3P56
 
 | |
3D00
 
 | |
4FM9
 
 | | Human topoisomerase II alpha bound to DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), DNA topoisomerase 2-alpha, ... | | Authors: | Wendorff, T.J, Schmidt, B.H, Heslop, P, Austin, C.A, Berger, J.M. | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Structure of DNA-Bound Human Topoisomerase II Alpha: Conformational Mechanisms for Coordinating Inter-Subunit Interactions with DNA Cleavage.
J.Mol.Biol., 424, 2012
|
|
4P7O
 
 | | Structure of Escherichia coli PgaB C-terminal domain, P1 crystal form | | Descriptor: | Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3H7W
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS017 | | Descriptor: | 2-nitro-N-(thiophen-3-ylmethyl)-4-(trifluoromethyl)aniline, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Key, J.M, Scheuermann, T.H, Anderson, P.C, Daggett, V, Gardner, K.H. | | Deposit date: | 2009-04-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B
J.Am.Chem.Soc., 131, 2009
|
|
1J47
 
 | |
3H82
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS020 | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(furan-2-ylmethyl)-2-nitro-4-(trifluoromethyl)aniline | | Authors: | Key, J.M, Scheuermann, T.H, Anderson, P.C, Daggett, V, Gardner, K.H. | | Deposit date: | 2009-04-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B
J.Am.Chem.Soc., 131, 2009
|
|
1M1P
 
 | | P21 crystal structure of the tetraheme cytochrome c3 from Shewanella oneidensis MR1 | | Descriptor: | HEME C, SULFATE ION, Small tetraheme cytochrome c | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
2E9R
 
 | | Foot-and-mouth disease virus RNA-dependent RNA polymerase in complex with a template-primer RNA and with ribavirin | | Descriptor: | 5'-R(*CP*AP*UP*GP*GP*GP*CP*CP*C)-3', 5'-R(*CP*CP*C*GP*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4P7Q
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7R
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with a poly-beta-1,6-N-acetyl-D-glucosamine (PNAG) hexamer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7N
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1MHL
 
 | | CRYSTAL STRUCTURE OF HUMAN MYELOPEROXIDASE ISOFORM C CRYSTALLIZED IN SPACE GROUP P2(1) AT PH 5.5 AND 20 DEG C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fenna, R.E, Zeng, J, Davey, C. | | Deposit date: | 1995-06-09 | | Release date: | 1996-01-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the green heme in myeloperoxidase.
Arch.Biochem.Biophys., 316, 1995
|
|
