8B39
 
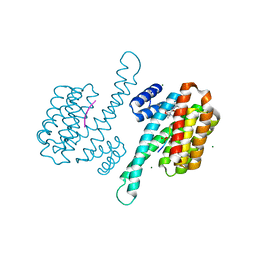 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080299) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[(2~{S},6~{R})-4-[(4-chlorophenyl)amino]-2,6-dimethyl-oxan-4-yl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8BO1
 
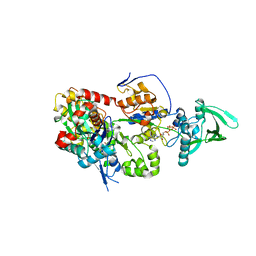 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AZIDE ION, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BBR
 
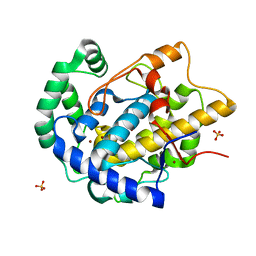 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, SULFATE ION | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
8BJI
 
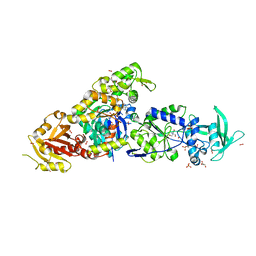 | | chimera of ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo fused to a proline-Rich-Domain (PRD) and profilin, bound to ADP-Mg-actin and a sulfate ion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BJE
 
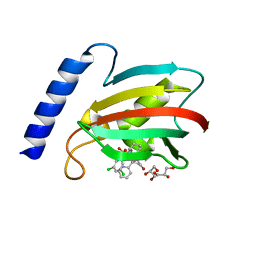 | | A structure of the truncated LpMIP with bound inhibitor JK236. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-[(1~{S})-1-pyridin-2-ylethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, GLYCEROL, Peptidyl-prolyl cis-trans isomerase | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BJC
 
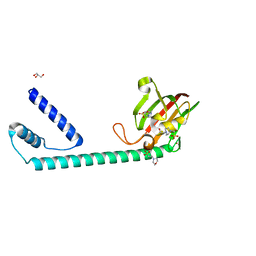 | | Full length structure of the apo-state LpMIP. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-03 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BR1
 
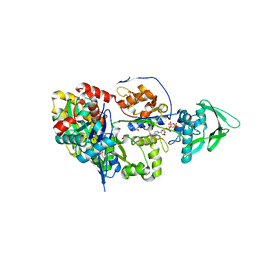 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-22 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
6WPR
 
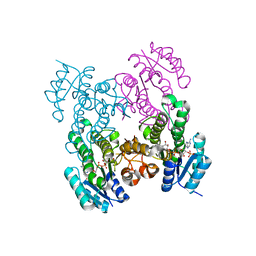 | |
8B53
 
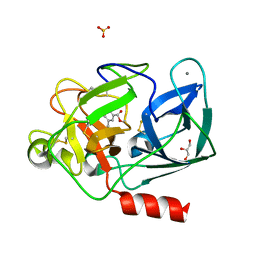 | |
6WQ8
 
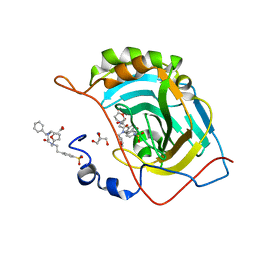 | | Carbonic Anhydrase II Complexed with 3-((2-((Furan-2-ylmethyl)(4-sulfamoylphenethyl)amino)-2-oxoethyl)(phenethyl)amino)propanoic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
8BRE
 
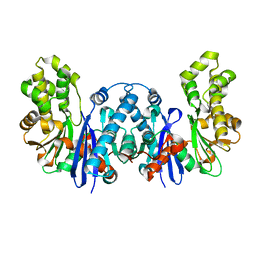 | |
8BJJ
 
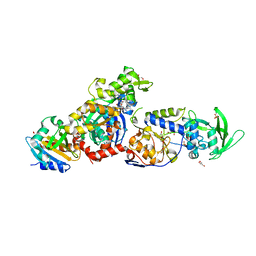 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to ATP-Mg-actin, human profilin 1 and a sulfate ion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
6WUP
 
 | |
8BR0
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin (residue Q3455 to L3863) in complex with 3'deoxyCTP and two manganese cations bound to Latrunculin-B-ADP-Mn-actin | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Texeira-Nuns, M, Retailleau, P, Renault, L. | | Deposit date: | 2022-11-22 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
6WT4
 
 | | Structure of a bacterial STING receptor from Flavobacteriaceae sp. in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Bacterial STING, SULFATE ION | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6WT7
 
 | | Structure of a metazoan TIR-STING receptor from C. gigas in complex with 2',3'-cGAMP | | Descriptor: | Metazoan TIR-STING fusion, cGAMP | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6WZX
 
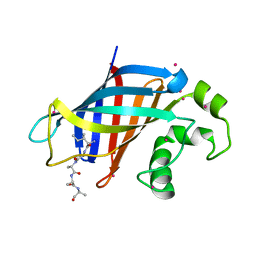 | | GID4 in complex with IGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, ILE-GLY-LEU-TRP-LYS peptide, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X0I
 
 | |
8B4K
 
 | |
6X1P
 
 | |
8B4J
 
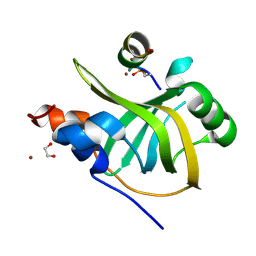 | | Rfa1-N-terminal domain in complex with phosphorylated Ddc2 | | Descriptor: | 1,2-ETHANEDIOL, DNA damage checkpoint protein LCD1, Replication factor A protein 1, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A DNA damage-induced phosphorylation circuit enhances Mec1 ATR Ddc2 ATRIP recruitment to Replication Protein A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6WYD
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-12 (AKA; 7-benzyl-1H-[1,2,3]triazolo[4,5-b]pyrid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-benzyl-1H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6X3O
 
 | | Co-structure of BTK kinase domain with L-005191930 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxo-3,5,6,7,8,8a-hexahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-methoxy-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8BHX
 
 | |
8BPT
 
 | |
