5AR0
 
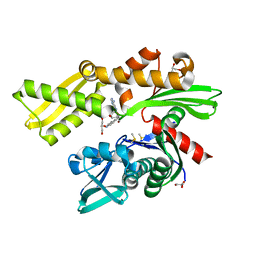 | | HSP72 with adenosine-derived inhibitor | | Descriptor: | (2R,3R,4S,5R)-2-(6-amino-8-((quinolin-7-ylmethyl)amino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Cheeseman, M.D, Westwood, I.M, Barbeau, O, Rowlands, M.G, Jones, A.M, Jeganathan, F, Burke, R, Dobson, S.E, Workman, P, Collins, I, van Montfort, R.L.M, Jones, K. | | Deposit date: | 2015-09-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploiting Protein Conformational Change to Optimize Adenosine-Derived Inhibitors of Hsp70.
J.Med.Chem., 59, 2016
|
|
5ABQ
 
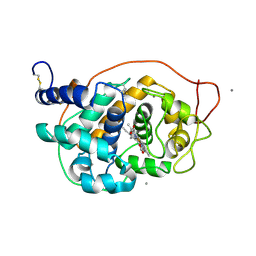 | | CRYSTAL STRUCTURE ANALYSIS OF FUNGAL VERSATILE PEROXIDASE FROM PLEUROTUS ERYNGII. MUTANT VPi-SS. MUTATED RESIDUES T2K, A49C, A61C, D69S, T70D, S86E, A131K, D146T, Q202L, Q219K, H232E, Q239R, L288R, S301K, A308R,A309K AND A314R. | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, VERSATILE PEROXIDASE | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2015-08-07 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Improving the Ph-Stability of Versatile Peroxidase by Comparative Structural Analysis with a Naturally-Stable Manganese Peroxidase.
Plos One, 10, 2015
|
|
5LMB
 
 | |
6RJ5
 
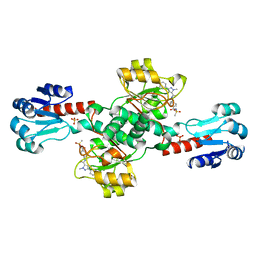 | | Crystal structure of PHGDH in complex with compound 39 | | Descriptor: | 2-methyl-~{N}-[(1~{R})-1-[4-(methylsulfonylcarbamoyl)phenyl]ethyl]-5-phenyl-pyrazole-3-carboxamide, D-3-phosphoglycerate dehydrogenase, SULFATE ION | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
7K9N
 
 | | Co-crystal structure of alpha glucosidase with compound 2 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(9-methoxynonyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7K9O
 
 | | Co-crystal structure of alpha glucosidase with compound 3 | | Descriptor: | (1~{S},2~{S},3~{R},4~{S},5~{S})-1-(hydroxymethyl)-5-[6-[[2-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]-4-(1,2,3,4-tetrazol-1-yl)phenyl]amino]hexylamino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7K9T
 
 | | Co-crystal structure of alpha glucosidase with compound 5 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-{[(5Z)-6-{[2-nitro-4-(2H-1,2,3-triazol-2-yl)phenyl]amino}hex-5-en-1-yl]amino}cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7K9Q
 
 | | Co-crystal structure of alpha glucosidase with compound 4 | | Descriptor: | (1S,2S,3R,4S,5S)-5-amino-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7KB6
 
 | | Co-crystal structure of alpha glucosidase with compound 7 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(6-{[2-nitro-4-(pyrimidin-2-yl)phenyl]amino}hexyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-10-01 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7KAD
 
 | | Co-crystal structure of alpha glucosidase with compound 6 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(6-{[2-nitro-4-(1H-1,2,3-triazol-1-yl)phenyl]amino}hexyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7KB8
 
 | | Co-crystal structure of alpha glucosidase with compound 8 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(4-{[2-nitro-4-(triazan-1-yl)phenyl]amino}butyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-10-01 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
4XRZ
 
 | | Human Cytochrome P450 2D6 BACE1 Inhibitor 6 Complex | | Descriptor: | (4aR,6R,8aS)-8a-(2,4-difluorophenyl)-6-(1H-pyrazol-4-yl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Cytochrome P450 2D6, GLYCEROL, ... | | Authors: | Johnson, E.F, Fan, Y. | | Deposit date: | 2015-01-21 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Utilizing Structures of CYP2D6 and BACE1 Complexes To Reduce Risk of Drug-Drug Interactions with a Novel Series of Centrally Efficacious BACE1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
2UZV
 
 | | PKA structures of indazole-pyridine series of AKT inhibitors | | Descriptor: | (2S)-1-[3-(CYCLOHEXYLMETHOXY)PHENYL]-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA,, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
7KPU
 
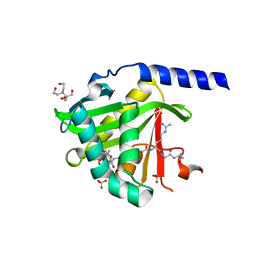 | |
7KNN
 
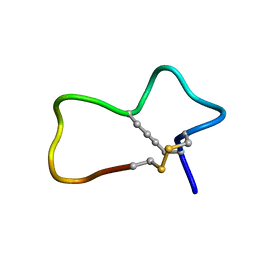 | |
5DGJ
 
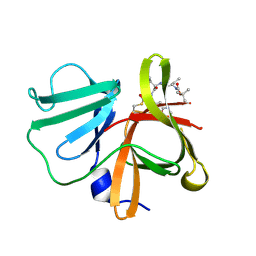 | | 1.0A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (20-mer) inhibitor | | Descriptor: | 3C-LIKE PROTEASE, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-22-oxa-6,9,14,20,21-pentaazabicyclo[17.2.1]docosa-1(21),19-dien-4-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-08-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
7JS8
 
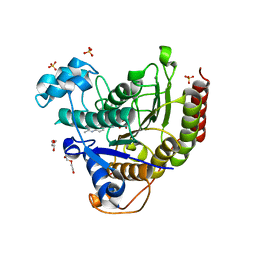 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH AN ETHYL KETONE INHIBITOR CONTAINING A SPIRO-BICYCLIC GROUP (COMPOUND 22) | | Descriptor: | (1S)-N-{(1S)-7,7-dihydroxy-1-[4-(2-methylquinolin-6-yl)-1H-imidazol-2-yl]nonyl}-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-08-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | Discovery of Ethyl Ketone-Based Highly Selective HDACs 1, 2, 3 Inhibitors for HIV Latency Reactivation with Minimum Cellular Potency Serum Shift and Reduced hERG Activity.
J.Med.Chem., 64, 2021
|
|
6T28
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6T29
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 18 (CS587) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3,5-bis(2-cyanopropan-2-yl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
7SB7
 
 | | Crystal structure of T. brucei hypoxanthine guanine phosphoribosyltransferase in complex with (4S,7S)-7-hydroxy-4-((guanin-9-yl)methyl)-2,5-dioxaheptan-1,7-diphosphonate | | Descriptor: | ({(2S)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2-[(2S)-2-hydroxy-2-phosphonoethoxy]propoxy}methyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Guddat, L.W, Keough, D.T. | | Deposit date: | 2021-09-24 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.64716625 Å) | | Cite: | Stereo-Defined Acyclic Nucleoside Phosphonates are Selective and Potent Inhibitors of Parasite 6-Oxopurine Phosphoribosyltransferases.
J.Med.Chem., 65, 2022
|
|
7SCR
 
 | | Crystal structure of trypanosome brucei hypoxanthine-guanine-xanthine phosphoribzosyltransferase in complex with (4S,7S)-7-hydroxy-4-((guanin-9-yl)methyl)-2,5-dioxaheptan-1,7-diphosphonate | | Descriptor: | ({(2S)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2-[(2S)-2-hydroxy-2-phosphonoethoxy]propoxy}methyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Guddat, L.W, Keough, D.T. | | Deposit date: | 2021-09-29 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12068486 Å) | | Cite: | Stereo-Defined Acyclic Nucleoside Phosphonates are Selective and Potent Inhibitors of Parasite 6-Oxopurine Phosphoribosyltransferases.
J.Med.Chem., 65, 2022
|
|
7RWI
 
 | | Mycobacterium tuberculosis RNA polymerase sigma L holoenzyme open promoter complex containing TNP-2198 | | Descriptor: | (3aM,9S,10bP,14S,15R,16S,17R,18R,19R,20S,21S,25R)-6,18,20-trihydroxy-14-methoxy-7,9,15,17,19,21,25-heptamethyl-1'-[2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl]-5,10,26-trioxo-3,5,9,10-tetrahydrospiro[9,4-(epoxypentadecanoimino)furo[2',3':7,8]naphtho[1,2-d]imidazole-2,4'-piperidin]-16-yl acetate, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design, Synthesis, and Characterization of TNP-2198, a Dual-Targeted Rifamycin-Nitroimidazole Conjugate with Potent Activity against Microaerophilic and Anaerobic Bacterial Pathogens.
J.Med.Chem., 65, 2022
|
|
5YOV
 
 | | Crystal structure of BRD4-BD1 bound with hjp126 | | Descriptor: | (3~{R})-4-cyclopentyl-~{N}-(2,4-dimethylphenyl)-1,3-dimethyl-2-oxidanylidene-3~{H}-quinoxaline-6-carboxamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Hu, J, Li, Y, Cao, D. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structure-based optimization of a series of selective BET inhibitors containing aniline or indoline groups.
Eur.J.Med.Chem., 150, 2018
|
|
5DG6
 
 | | 2.35A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (21-mer) inhibitor | | Descriptor: | 3C-LIKE PROTEASE, CHLORIDE ION, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-23-oxa-6,9,14,21,22-pentaazabicyclo[18.2.1]tricosa-1(22),20-dien-4-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-08-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
6T5B
 
 | | KRasG12C ligand complex | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Phillips, C. | | Deposit date: | 2019-10-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
