5XV7
 
 | | SRPK1 in complex with Alectinib | | Descriptor: | 1,2-ETHANEDIOL, 9-ethyl-6,6-dimethyl-8-[4-(morpholin-4-yl)piperidin-1-yl]-11-oxo-6,11-dihydro-5H-benzo[b]carbazole-3-carbonitrile, serine-arginine (SR) protein kinase 1 | | Authors: | Zeng, C, Ngo, J.C.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | SRPKIN-1: A Covalent SRPK1/2 Inhibitor that Potently Converts VEGF from Pro-angiogenic to Anti-angiogenic Isoform
Cell Chem Biol, 25, 2018
|
|
5CF8
 
 | | CRYSTAL STRUCTURE OF JANUS KINASE 2 IN COMPLEX WITH N,N-DICYCLOPROPYL-10-ETHYL-7-[(3-METHOXYPROPYL)AMINO] -3-METHYL-3,5,8,10-TETRAAZATRICYCLO[7.3.0.0,6] DODECA-1(9),2(6),4,7,11-PENTAENE-11-CARBOXAMIDE | | Descriptor: | N,N-dicyclopropyl-4-[(1,5-dimethyl-1H-pyrazol-3-yl)amino]-6-ethyl-1-methyl-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Sack, J.S. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Highly Selective JAK2 Inhibitor, BMS-911543, for the Treatment of Myeloproliferative Neoplasms.
Acs Med.Chem.Lett., 6, 2015
|
|
6J3F
 
 | | Crystal structure of the glutathione S-transferase, CsGST63524, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a serine-type glutathione S-transferase of Ceriporiopsis subvermispora and identification of the enzymatically important non-canonical residues by functional mutagenesis.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
1K9W
 
 | | HIV-1(MAL) RNA Dimerization Initiation Site | | Descriptor: | HIV-1 DIS(Mal)UU RNA, MAGNESIUM ION | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site.
Nat.Struct.Biol., 8, 2001
|
|
5CDO
 
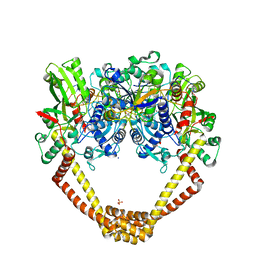 | | 3.15A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-4',6'-dihydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidin]-2'-one, (2R,4S,4aS,5R)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, (2R,4S,4aS,5S)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
6IZ4
 
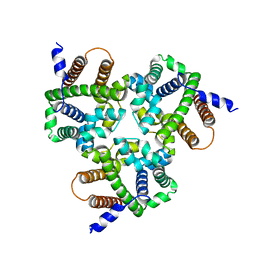 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | Trimeric intracellular cation channel type B-B | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5CDP
 
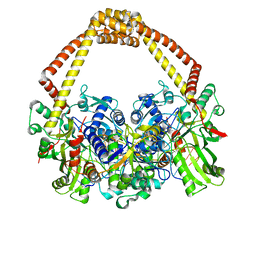 | | 2.45A structure of etoposide with S.aureus DNA gyrase and DNA | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
3JTG
 
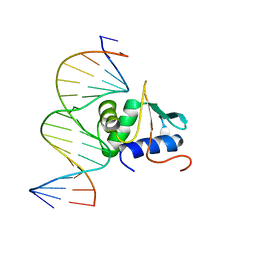 | | Crystal structure of mouse Elf3 C-terminal DNA-binding domain in complex with type II TGF-beta receptor promoter DNA | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*CP*AP*GP*GP*AP*AP*AP*CP*TP*CP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*GP*TP*TP*TP*CP*CP*TP*GP*TP*TP*T)-3'), ETS-related transcription factor Elf-3 | | Authors: | Tahirov, T.H, Babayeva, N.D, Agarkar, V.B, Rizzino, A. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of mouse Elf3 C-terminal DNA-binding domain in complex with type II TGF-beta receptor promoter DNA.
J.Mol.Biol., 397, 2010
|
|
5TBR
 
 | |
5GN7
 
 | | Crystal structure of alternative oxidase from Trypanosoma brucei brucei complexed with cumarin derivative-17 | | Descriptor: | 4-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-7,8-bis(oxidanyl)chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5KBR
 
 | | Pak1 in complex with 7-azaindole inhibitor | | Descriptor: | (4-chlorophenyl)-[5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]methanone, Serine/threonine-protein kinase PAK 1 | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Optimization of Highly Kinase Selective Bis-anilino Pyrimidine PAK1 Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
6OKT
 
 | |
7W0I
 
 | |
5IEK
 
 | |
6BL8
 
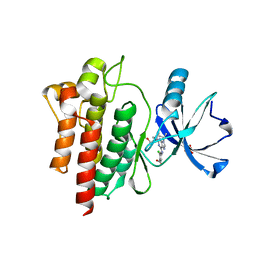 | |
2VWJ
 
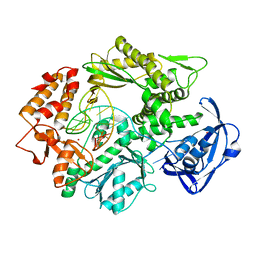 | | Uracil Recognition in Archaeal DNA Polymerases Captured by X-ray Crystallography. | | Descriptor: | 5'-D(*AP*AP*UP*GP*GP*AP*GP*AP*CP*GP *GP*CP*TP*TP*TP*TP*GP*CP*CP*GP*TP*GP*TP*C)-3', DNA POLYMERASE, POTASSIUM ION | | Authors: | Firbank, S.J, Wardle, J, Heslop, P, Lewis, R.J, Connolly, B.A. | | Deposit date: | 2008-06-25 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Uracil Recognition in Archaeal DNA Polymerases Captured by X-Ray Crystallography.
J.Mol.Biol., 381, 2008
|
|
1GFZ
 
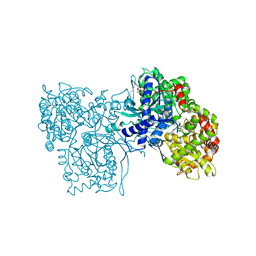 | | FLAVOPIRIDOL INHIBITS GLYCOGEN PHOSPHORYLASE BY BINDING AT THE INHIBITOR SITE | | Descriptor: | CAFFEINE, GLYCOGEN PHOSPHORYLASE, INOSINIC ACID, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Tsitsanou, K.E, Johnson, L.N. | | Deposit date: | 2000-06-29 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Flavopiridol inhibits glycogen phosphorylase by binding at the inhibitor site.
J.Biol.Chem., 275, 2000
|
|
6MTO
 
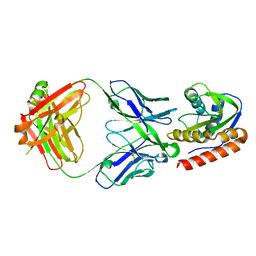 | | Crystal structure of VRC42.01 Fab in complex with T117-F MPER scaffold | | Descriptor: | Antibody VRC42.01 Fab heavy chain, Antibody VRC42.01 Fab light chain, VRC42 epitope T117-F scaffold | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MTR
 
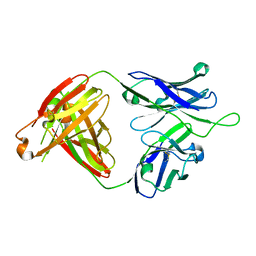 | | Crystal structure of VRC43.01 Fab | | Descriptor: | Antibody VRC43.01 Fab heavy chain, Antibody VRC43.01 Fab light chain | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6FNL
 
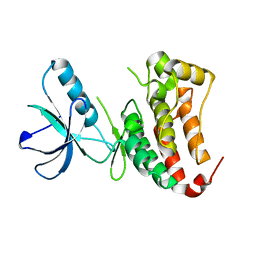 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase | | Descriptor: | Ephrin type-B receptor 4 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
4WOJ
 
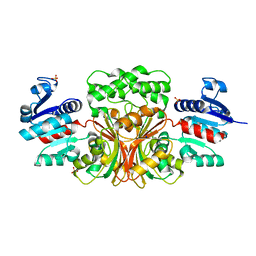 | | Aspartate Semialdehyde Dehydrogenase from Francisella tularensis | | Descriptor: | Aspartate semialdehyde dehydrogenase, SODIUM ION, SULFATE ION | | Authors: | Mank, N.J, Arnette, A.K, Klapper, V.G, Chruszcz, M. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of aspartate b-semialdehyde dehydrogenase from Francisella tularensis
Acta Crystallogr.,Sect.F, 74, 2018
|
|
6FNI
 
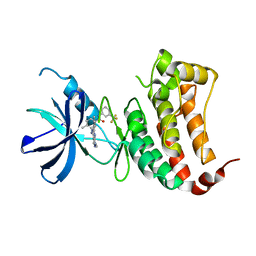 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase with NVP-BHG712 | | Descriptor: | 4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-B receptor 4 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
5T4Z
 
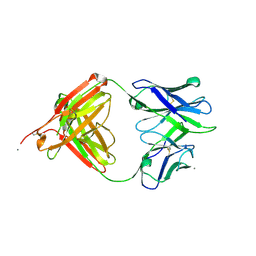 | |
6OKU
 
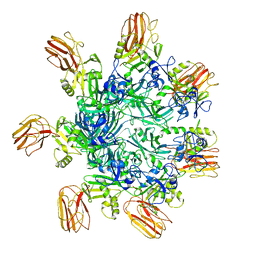 | |
6VN7
 
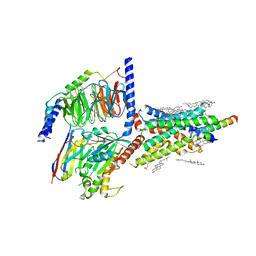 | | Cryo-EM structure of an activated VIP1 receptor-G protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Duan, J, Shen, D.-D, Zhou, X.E, Liu, Q.-F, Zhuang, Y.-W, Zhang, H.-B, Xu, P.-Y, Ma, S.-S, He, X.-H, Melcher, K, Zhang, Y, Xu, H.E, Yi, J. | | Deposit date: | 2020-01-29 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy.
Nat Commun, 11, 2020
|
|
