5SVR
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel bound to competitive antagonist A-317491 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-{[(3-phenoxyphenyl)methyl][(1S)-1,2,3,4-tetrahydronaphthalen-1-yl]carbamoyl}benzene-1,2,4-tricarboxylic acid, MAGNESIUM ION, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-07 | | Release date: | 2016-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5UIP
 
 | | structure of DHFR with bound DAP, p-ABG and NADP | | Descriptor: | Dihydrofolate reductase, N-(4-aminobenzene-1-carbonyl)-L-glutamic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pedersen, L.C, London, R.E. | | Deposit date: | 2017-01-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Structural Basis for Biguanide Activity.
Biochemistry, 56, 2017
|
|
5V4I
 
 | | Osmium(II)(cymene)(chlorido)2-lysozyme adduct with one binding site | | Descriptor: | Lysozyme C, SODIUM ION, dichloro[(1,2,3,4,5,6-eta)-3-methyl-6-(propan-2-yl)benzene-1,2,4,5-tetrayl]osmium | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The metalation of hen egg white lysozyme impacts protein stability as shown by ion mobility mass spectrometry, differential scanning calorimetry, and X-ray crystallography.
Chem. Commun. (Camb.), 53, 2017
|
|
5SA7
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1673618163 | | Descriptor: | 4-amino-N-(2-hydroxyethyl)-N-methylbenzene-1-sulfonamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5UIO
 
 | |
5UKF
 
 | | Crystal Structure of the Human Vaccinia-related Kinase 1 Bound to an Oxindole Inhibitor | | Descriptor: | 4-{[(Z)-(7-oxo-6,7-dihydro-8H-[1,3]thiazolo[5,4-e]indol-8-ylidene)methyl]amino}benzene-1-sulfonamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Counago, R.M, Wells, C, Zuercher, W, Willson, T.M, Bountra, C, Edwards, A.M, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-22 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations.
Sci Rep, 7, 2017
|
|
5V2O
 
 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | Descriptor: | 1,3,5-BENZENETRICARBOXYLIC ACID, GLYCEROL, NONAETHYLENE GLYCOL, ... | | Authors: | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | Deposit date: | 2017-03-06 | | Release date: | 2017-10-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U41
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 16 | | Descriptor: | 6-[2-({benzyl[4-(thiophen-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, POTASSIUM ION, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2Z9B
 
 | |
5U3R
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 2 | | Descriptor: | 6-[2-({[4-(furan-2-yl)benzene-1-carbonyl](propan-2-yl)amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U3Z
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 10 | | Descriptor: | 6-[2-({cyclopropyl[4-(furan-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U3V
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 6 | | Descriptor: | 6-[2-({cyclopentyl[4-(furan-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UUZ
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P200 | | Descriptor: | 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-17 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P200
To Be Published
|
|
5V8T
 
 | | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354 | | Descriptor: | 2-{[3,5-bis(2-methoxyethoxy)benzene-1-carbonyl]amino}ethyl (2S)-1-(benzylsulfonyl)piperidine-2-carboxylate, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Lorimer, D.D, Dranow, D.M, Seufert, F, Abendroth, J, Holzgrabe, U. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354
to be published
|
|
5UPZ
 
 | | HIV-1 wild Type protease with GRL-0518A , an isophthalamide-derived P2-P3 ligand with the para-hydoxymethyl sulfonamide isostere as the P2' group | | Descriptor: | CHLORIDE ION, GLYCEROL, N~3~-{(2S,3R)-3-hydroxy-4-[{[4-(hydroxymethyl)phenyl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}-N~1~-methyl-N~1~-[(4-methyl-1,3-oxazol-2-yl)methyl]benzene-1,3-dicarboxamide, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Design of novel HIV-1 protease inhibitors incorporating isophthalamide-derived P2-P3 ligands: Synthesis, biological evaluation and X-ray structural studies of inhibitor-HIV-1 protease complex.
Bioorg. Med. Chem., 25, 2017
|
|
5T8H
 
 | |
5UZS
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P200 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-22 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P200
To Be Published
|
|
5R94
 
 | |
2A32
 
 | | Trypsin in complex with benzene boronic acid | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, BORATE ION, CALCIUM ION, ... | | Authors: | Transue, T.R, Gabel, S.A, London, R.E. | | Deposit date: | 2005-06-23 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NMR and crystallographic characterization of adventitious borate binding by trypsin.
Bioconjug.Chem., 17, 2006
|
|
5V5N
 
 | | Crystal structure of Takinib bound to TAK1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7/TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, N~1~-(1-propyl-1,3-dihydro-2H-benzimidazol-2-ylidene)benzene-1,3-dicarboxamide | | Authors: | Gurbani, D, Westover, K, Bera, A.K. | | Deposit date: | 2017-03-14 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Takinib, a Selective TAK1 Inhibitor, Broadens the Therapeutic Efficacy of TNF-alpha Inhibition for Cancer and Autoimmune Disease.
Cell Chem Biol, 24, 2017
|
|
5VIO
 
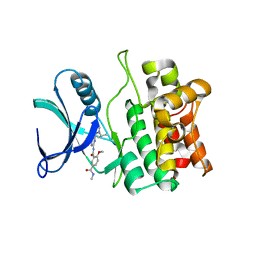 | | Crystal structure of ASK1 kinase domain with a potent inhibitor (analog 13) | | Descriptor: | 4-methoxy-N~1~-methyl-N~3~-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}benzene-1,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Jasti, J, Chang, J, Kurumbail, R. | | Deposit date: | 2017-04-17 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Rational approach to highly potent and selective apoptosis signal-regulating kinase 1 (ASK1) inhibitors.
Eur J Med Chem, 145, 2017
|
|
2DEF
 
 | |
7MF0
 
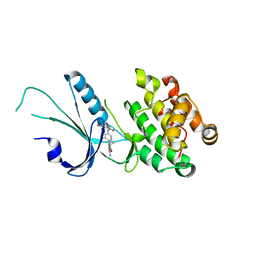 | | Co-crystal structure of PERK with inhibitor (R)-2-amino-N-cyclopropyl-5-(4-(2-(3,5-difluorophenyl)-2-hydroxyacetamido)-2-methylphenyl)nicotinamide | | Descriptor: | 2-amino-N-cyclopropyl-5-(4-{[(2R)-2-(3,5-difluorophenyl)-2-hydroxyacetyl]amino}-2-methylphenyl)pyridine-3-carboxamide, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wiens, B, Koszelak-Rosenblum, M, Surman, M.D, Zhu, G, Mulvihill, M.J. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Discovery of 2-amino-3-amido-5-aryl-pyridines as highly potent, orally bioavailable, and efficacious PERK kinase inhibitors.
Bioorg.Med.Chem.Lett., 43, 2021
|
|
7ML7
 
 | |
7MFR
 
 | | Crystal Structure of a Fab fragment bound to peptide GGM | | Descriptor: | Antibody fragment - Heavy Chain of fab, Antibody fragment - Light Chain of fab, GLY-GLY-MET, ... | | Authors: | Sudhamsu, J. | | Deposit date: | 2021-04-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.848 Å) | | Cite: | Antibody toolkit reveals N-terminally ubiquitinated substrates of UBE2W.
Nat Commun, 12, 2021
|
|
