6YME
 
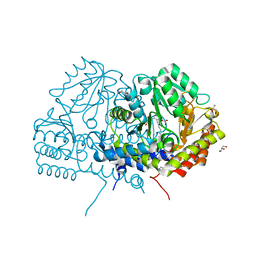 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-internal aldimine state | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Serine hydroxymethyltransferase | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YMD
 
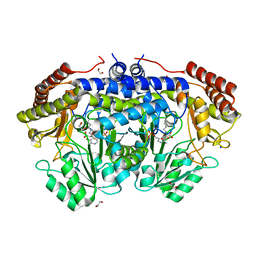 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the covalent complex with malonate | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, MALONATE ION, ... | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
5T99
 
 | |
5TA5
 
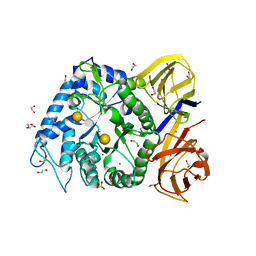 | | Crystal structure of BuGH86wt in complex with neoagarooctaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5T9X
 
 | | Crystal structure of BuGH16Bwt | | Descriptor: | Glycoside Hydrolase, IMIDAZOLE, SODIUM ION | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
7A1R
 
 | |
4L7K
 
 | | Crystal Structure of Ketosteroid Isomerase D38E from Pseudomonas Testosteroni (tKSI) | | Descriptor: | GLYCEROL, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2013-06-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Use of anion-aromatic interactions to position the general base in the ketosteroid isomerase active site.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5T98
 
 | | Crystal structure of BuGH2Awt | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside Hydrolase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5TA1
 
 | | Crystal structure of BuGH86wt | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5TA9
 
 | |
4Z5T
 
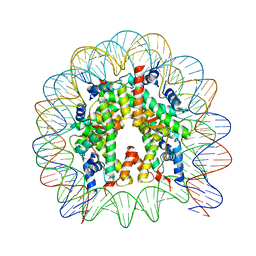 | | The nucleosome containing human H3.5 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Urahama, T, Harada, A, Maehara, K, Horikoshi, N, Sato, K, Sato, Y, Shiraishi, K, Sugino, N, Osakabe, A, Tachiwana, H, Kagawa, W, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-04-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Histone H3.5 forms an unstable nucleosome and accumulates around transcription start sites in human testis.
Epigenetics Chromatin, 9, 2016
|
|
5T9A
 
 | | Crystal structure of BuGH2Cwt | | Descriptor: | 1,2-ETHANEDIOL, Glycoside Hydrolase, SULFATE ION | | Authors: | Pluvinage, B, Boraston, A.B, Abbott, W.D. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5TA0
 
 | | Crystal structure of BuGH86E322Q in complex with neoagarooctaose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Boraston, A.B, Abbott, W.D. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5T9G
 
 | |
5TA7
 
 | | Crystal structure of BuGH117Bwt | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside Hydrolase, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5X57
 
 | | Structure of GAR domain of ACF7 | | Descriptor: | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, NICKEL (II) ION | | Authors: | Yang, F, Wang, T, Zhang, Y, Wu, X.Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | ACF7 regulates inflammatory colitis and intestinal wound response by orchestrating tight junction dynamics.
Nat Commun, 8, 2017
|
|
1T5Z
 
 | | Crystal Structure of the Androgen Receptor Ligand Binding Domain (LBD) with DHT and a peptide derived from its physiological coactivator ARA70 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 4 | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-05-05 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor.
J.Biol.Chem., 280, 2005
|
|
1KEY
 
 | | Crystal Structure of Mouse Testis/Brain RNA-binding Protein (TB-RBP) | | Descriptor: | translin | | Authors: | Pascal, J.M, Hart, P.J, Hecht, N.B, Robertus, J.D. | | Deposit date: | 2001-11-19 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of TB-RBP, a Novel RNA-binding and Regulating Protein
J.Mol.Biol., 319, 2002
|
|
6YMF
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-Serine external aldimine state | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Serine hydroxymethyltransferase, ... | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
7ZW8
 
 | |
7ZY6
 
 | |
6TRO
 
 | | Crystal structure of the T-cell receptor GVY01 bound to HLA A2*01-GVYDGREHTV | | Descriptor: | Beta-2-microglobulin, MAGE-A4 peptide (amino acids 230-239), MHC class I antigen, ... | | Authors: | Coles, C.H, McMurran, C, Lloyd, A, Hibbert, L, Lupardus, P.J, Cole, D.K, Harper, S. | | Deposit date: | 2019-12-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | T cell receptor interactions with human leukocyte antigen govern indirect peptide selectivity for the cancer testis antigen MAGE-A4.
J.Biol.Chem., 295, 2020
|
|
6TRN
 
 | | Crystal structure of HLA A2*01-AVYDGREHTV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MAGE-A4 peptide (amino acids 230-239) variant, ... | | Authors: | Coles, C.H, McMurran, C, Lloyd, A, Hibbert, L, Lupardus, P.J, Cole, D.K, Harper, S. | | Deposit date: | 2019-12-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | T cell receptor interactions with human leukocyte antigen govern indirect peptide selectivity for the cancer testis antigen MAGE-A4.
J.Biol.Chem., 295, 2020
|
|
5WBS
 
 | | Crystal structure of Frizzled-7 CRD with an inhibitor peptide Fz7-21 | | Descriptor: | Frizzled-7,inhibitor peptide Fz7-21 | | Authors: | Nile, A.H, Mukund, S, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
3AFA
 
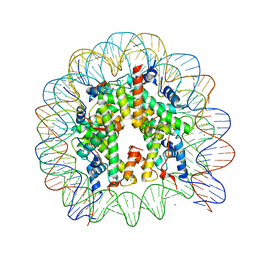 | | The human nucleosome structure | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Tachiwana, H, Kagawa, W, Osakabe, A, Koichiro, K, Shiga, T, Kimura, H, Kurumizaka, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
