2F10
 
 | | Crystal Structure of the Human Sialidase Neu2 in Complex with Peramivir inhibitor | | Descriptor: | 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, McKimm-Breschkin, J, Colman, P.M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with Peramivir inhibitor
To be Published
|
|
2F11
 
 | | Crystal Structure of the Human Sialidase Neu2 in Complex with isobutyl ether mimetic Inhibitor | | Descriptor: | 2-methylpropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with isobutyl ether mimetic Inhibitor
To be Published
|
|
2F12
 
 | | Crystal Structure of the Human Sialidase Neu2 in Complex with 3- hydroxypropyl ether mimetic Inhibitor | | Descriptor: | 3-hydroxypropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with 3- hydroxypropyl ether mimetic Inhibitor
To be Published
|
|
2F13
 
 | | Crystal Structure of the Human Sialidase Neu2 in Complex with 2',3'- dihydroxypropyl ether mimetic Inhibitor | | Descriptor: | (2R)-2,3-dihydroxypropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with 2',3'- dihydroxypropyl ether mimetic Inhibitor
To be Published
|
|
2F14
 
 | | Tne Crystal Structure of the Human Carbonic Anhydrase II in Complex with a Fluorescent Inhibitor | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, 5-{[({2-[4-(AMINOSULFONYL)PHENYL]ETHYL}AMINO)CARBONOTHIOYL]AMINO}-2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Alterio, V, Pedone, C, De Simone, G. | | Deposit date: | 2005-11-14 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray and molecular modeling study for the interaction of a fluorescent antitumor sulfonamide with isozyme II and IX.
J.Am.Chem.Soc., 128, 2006
|
|
2F15
 
 | | Glycogen-Binding Domain Of The Amp-Activated Protein Kinase beta2 Subunit | | Descriptor: | 5'-AMP-activated protein kinase, beta-2 subunit | | Authors: | Walker, J.R, Wybenga-Groot, L, Finerty Jr, P.J, Newman, E, MacKenzie, F.M, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-14 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Glycogen-Binding Domain Of The Amp-Activated Protein Kinase beta2 Subunit
To be Published
|
|
2F16
 
 | | Crystal structure of the yeast 20S proteasome in complex with bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M. | | Deposit date: | 2005-11-14 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Boronic Acid-Based Proteasome Inhibitor Bortezomib in Complex with the Yeast 20S Proteasome.
Structure, 14, 2006
|
|
2F17
 
 | | Mouse Thiamin Pyrophosphokinase in a Ternary Complex with Pyrithiamin Pyrophosphate and AMP at 2.5 angstrom | | Descriptor: | 1-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-2-METHYLPYRIDINIUM, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Liu, J.Y, Timm, D.E, Hurley, T.D. | | Deposit date: | 2005-11-14 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyrithiamine as a substrate for thiamine pyrophosphokinase
J.Biol.Chem., 281, 2006
|
|
2F18
 
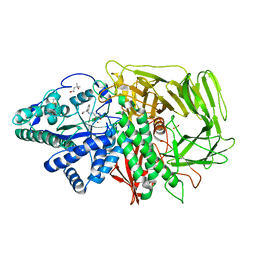 | | GOLGI ALPHA-MANNOSIDASE II complex with (2R,3R,4S)-2-({[(1R)-2-hydroxy-1-phenylethyl]amino}methyl)pyrrolidine-3,4-diol | | Descriptor: | (2R,3R,4S)-2-({[(1R)-2-HYDROXY-1-PHENYLETHYL]AMINO}METHYL)PYRROLIDINE-3,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evaluation of docking programs for predicting binding of Golgi alpha-mannosidase II inhibitors: a comparison with crystallography.
Proteins, 69, 2007
|
|
2F19
 
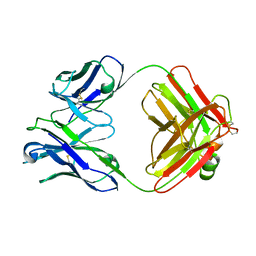 | | THREE-DIMENSIONAL STRUCTURE OF TWO CRYSTAL FORMS OF FAB R19.9, FROM A MONOCLONAL ANTI-ARSONATE ANTIBODY | | Descriptor: | IGG2B-KAPPA R19.9 FAB (HEAVY CHAIN), IGG2B-KAPPA R19.9 FAB (LIGHT CHAIN) | | Authors: | Lascombe, M.B, Alzari, P.M, Poljak, R.J, Nisonoff, A. | | Deposit date: | 1992-05-27 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of two crystal forms of FabR19.9 from a monoclonal anti-arsonate antibody.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
2F1A
 
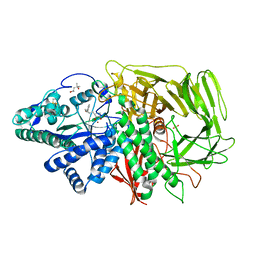 | | GOLGI ALPHA-MANNOSIDASE II COMPLEX WITH (2R,3R,4S)-2-({[(1S)-2-hydroxy-1-phenylethyl]amino}methyl)pyrrolidine-3,4-diol | | Descriptor: | (2R,3R,4S)-2-({[(1S)-2-HYDROXY-1-PHENYLETHYL]AMINO}METHYL)PYRROLIDINE-3,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Evaluation of docking programs for predicting binding of Golgi alpha-mannosidase II inhibitors: a comparison with crystallography.
Proteins, 69, 2007
|
|
2F1B
 
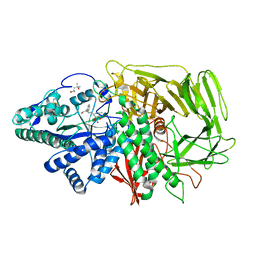 | | GOLGI ALPHA-MANNOSIDASE II COMPLEX WITH (2R,3R,4S,5R)-2-({[(1R)-2-hydroxy-1-phenylethyl]amino}methyl)-5-methylpyrrolidine-3,4-diol | | Descriptor: | (2R,3R,4S,5R)-2-({[(1R)-2-HYDROXY-1-PHENYLETHYL]AMINO}METHYL)-5-METHYLPYRROLIDINE-3,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Evaluation of docking programs for predicting binding of Golgi alpha-mannosidase II inhibitors: a comparison with crystallography.
Proteins, 69, 2007
|
|
2F1C
 
 | |
2F1D
 
 | | X-Ray Structure of imidazoleglycerol-phosphate dehydratase | | Descriptor: | Imidazoleglycerol-phosphate dehydratase 1, MANGANESE (II) ION, SULFATE ION | | Authors: | Rice, D.W, Glynn, S.E, Baker, P.J, Sedelnikova, S.E, Davies, C.L, Eadsforth, T.C. | | Deposit date: | 2005-11-14 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of imidazoleglycerol-phosphate dehydratase.
Structure, 13, 2005
|
|
2F1E
 
 | | Solution structure of ApaG protein | | Descriptor: | Protein apaG | | Authors: | Contessa, G, Pertinhez, T.A, Spisni, A, Paci, M, Farah, C.S, Cicero, D.O. | | Deposit date: | 2005-11-14 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ApaG from Xanthomonas axonopodis pv. citri reveals a fibronectin-3 fold.
Proteins, 67, 2007
|
|
2F1F
 
 | | Crystal structure of the regulatory subunit of acetohydroxyacid synthase isozyme III from E. coli | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Acetolactate synthase isozyme III small subunit, MAGNESIUM ION, ... | | Authors: | Kaplun, A, Vyazmensky, M, Barak, Z, Chipman, D.M, Shaanan, B. | | Deposit date: | 2005-11-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Regulatory Subunit of Acetohydroxyacid Synthase Isozyme III from Escherichia coli.
J.Mol.Biol., 357, 2006
|
|
2F1G
 
 | | Cathepsin S in complex with non-covalent 2-(Benzoxazol-2-ylamino)-acetamide | | Descriptor: | Cathepsin S, GLYCEROL, N~2~-1,3-BENZOXAZOL-2-YL-3-CYCLOHEXYL-N-{2-[(4-METHOXYPHENYL)AMINO]ETHYL}-L-ALANINAMIDE | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2005-11-14 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of arylaminoethyl amides as noncovalent inhibitors of cathepsin S. Part 3: Heterocyclic P3.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2F1H
 
 | | RECOMBINASE IN COMPLEX WITH AMP-PNP and Potassium | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Qian, X, He, Y, Wu, Y, Luo, Y. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae.
J.Mol.Biol., 360, 2006
|
|
2F1I
 
 | | Recombinase in Complex with AMP-PNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Qian, X, He, Y, Wu, Y, Luo, Y. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae.
J.Mol.Biol., 360, 2006
|
|
2F1J
 
 | | Recombinase in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION | | Authors: | Qian, X, He, Y, Wu, Y, Luo, Y. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae.
J.Mol.Biol., 360, 2006
|
|
2F1K
 
 | | Crystal structure of Synechocystis arogenate dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, prephenate dehydrogenase | | Authors: | Legrand, P, Dumas, R, Seux, M, Rippert, P, Ravelli, R, Ferrer, J.-L, Matringe, M. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical Characterization and Crystal Structure of Synechocystis Arogenate Dehydrogenase Provide Insights into Catalytic Reaction
Structure, 14, 2006
|
|
2F1L
 
 | |
2F1M
 
 | |
2F1N
 
 | |
2F1O
 
 | | Crystal Structure of NQO1 with Dicoumarol | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Shaul, Y, Asher, G, Dym, O, Tsvetkov, P, Adler, J, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-11-15 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of NAD(P)H quinone oxidoreductase 1 in complex with its potent inhibitor dicoumarol.
Biochemistry, 45, 2006
|
|
