3BBF
 
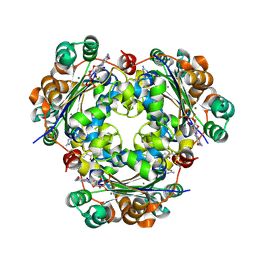 | | Crystal structure of the NM23-H2 transcription factor complex with GDP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Weichsel, A, Montfort, W.R. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NM23-H2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element III1.
Mol.Cancer Ther., 8, 2009
|
|
8Q5V
 
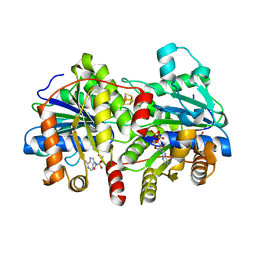 | |
6PBC
 
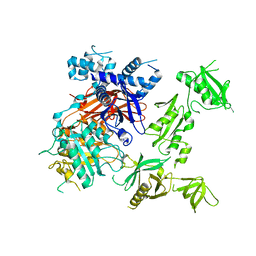 | | Structural basis for the activation of PLC-gamma isozymes by phosphorylation and cancer-associated mutations | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma,1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, CALCIUM ION, SODIUM ION | | Authors: | Hajicek, N, Sondek, J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the activation of PLC-gamma isozymes by phosphorylation and cancer-associated mutations.
Elife, 8, 2019
|
|
8XHM
 
 | | Crystal structure of Nocardia seriolae dUTP diphosphatase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Weng, J, Wei, Z. | | Deposit date: | 2023-12-18 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-guided discovery of novel dUTPase inhibitors with anti- Nocardia activity by computational design.
J Enzyme Inhib Med Chem, 39, 2024
|
|
6PHZ
 
 | | Crystal structure of Marinobacter subterrani acetylpolyamine amidohydrolase (msAPAH) complexed with 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptan-2-one | | Descriptor: | 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptane-2,2-diol, Acetylpolyamine Amidohydrolase, MAGNESIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-06-25 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of the Acetylpolyamine Amidohydrolase from the Deep Earth HalophileMarinobacter subterrani.
Biochemistry, 58, 2019
|
|
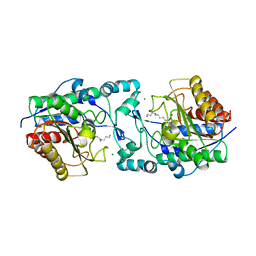
8BP6
 
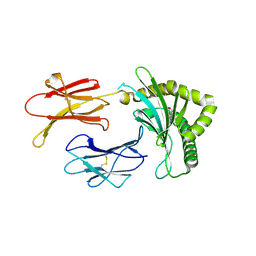 | | Structure of MHC-class I related molecule MR1 with bound M3Ade. | | Descriptor: | (1R,5S)-8-(9H-purin-6-yl)-2-oxa-8-azabicyclo[3.3.1]nona-3,6-diene-4,6-dicarbaldehyde, Beta-2-microglobulin,Major histocompatibility complex class I-related gene protein | | Authors: | Berloffa, G, Jakob, R.P, Maier, T. | | Deposit date: | 2022-11-16 | | Release date: | 2023-11-29 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The carbonyl nucleobase adduct M 3 Ade is a potent antigen for adaptive polyclonal MR1-restricted T cells.
Immunity, 2024
|
|
8PSR
 
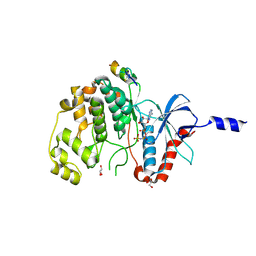 | | ERK2 covalently bound to SynthRevD-12-opt artificial peptide | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Gogl, G, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
3VDR
 
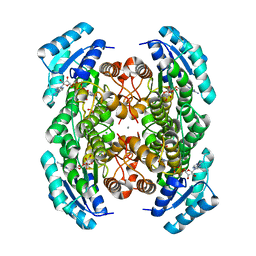 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase, prepared in the presence of the substrate D-3-hydroxybutyrate and NAD(+) | | Descriptor: | (3R)-3-hydroxybutanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETOACETIC ACID, ... | | Authors: | Hoque, M.M, Shimizu, S, Juan, E.C.M, Sato, Y, Hossain, M.T, Yamamoto, T, Imamura, S, Amano, H, Suzuki, K, Sekiguchi, T, Tsunoda, M, Takenaka, A. | | Deposit date: | 2012-01-06 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of D-3-hydroxybutyrate dehydrogenase prepared in the presence of the substrate D-3-hydroxybutyrate and NAD+.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2PJL
 
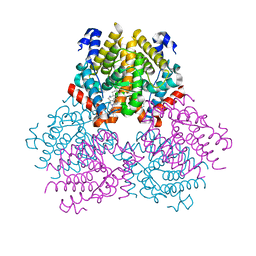 | |
9IL3
 
 | |
9HL0
 
 | | Protein Kinase CK2 and small molecule ligands | | Descriptor: | 5,7-bis(fluoranyl)-1~{H}-indole, Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krimm, I, Gelin, M, Guichou, J.F. | | Deposit date: | 2024-12-04 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Binding-Site Switch for Protein Kinase CK2 Inhibitors.
Chemmedchem, 20, 2025
|
|
8CJB
 
 | | A268M variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | ACETATE ION, CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coupling CO 2 Reduction and Acetyl-CoA Formation: The Role of a CO Capturing Tunnel in Enzymatic Catalysis.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1JDD
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 2) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
9HL7
 
 | | Protein Kinase CK2 and small molecule ligands | | Descriptor: | 2-(6-chloranyl-1~{H}-indol-3-yl)ethanoic acid, Casein kinase II subunit alpha, DIMETHYL SULFOXIDE, ... | | Authors: | Krimm, I, Gelin, I, Guichou, J.F. | | Deposit date: | 2024-12-04 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Binding-Site Switch for Protein Kinase CK2 Inhibitors.
Chemmedchem, 20, 2025
|
|
9HKS
 
 | |
3W1A
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with 5-halogenated orotate derivatives | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-13 | | Release date: | 2013-11-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with 5-halogenated orotate derivatives
To be Published
|
|
3W23
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-131 | | Descriptor: | 5-[2-(3-chlorophenyl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-131
To be Published
|
|
3VB5
 
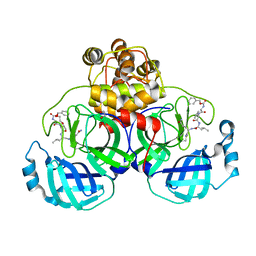 | | Crystal structure of SARS-CoV 3C-like protease with C4Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, C4Z inhibitor | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
6PKH
 
 | |
3BGL
 
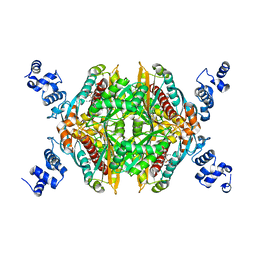 | | Hepatoselectivity of Statins: Design and synthesis of 4-sulfamoyl pyrroles as HMG-CoA reductase inhibitors | | Descriptor: | (3R,5R)-7-[2-(4-fluorophenyl)-5-(1-methylethyl)-4-(morpholin-4-ylsulfonyl)-3-phenyl-1H-pyrrol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Finzel, B.C, Pavlovsky, A, Park, W.K.C. | | Deposit date: | 2007-11-26 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | Hepatoselectivity of statins: design and synthesis of 4-sulfamoyl pyrroles as HMG-CoA reductase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2PIJ
 
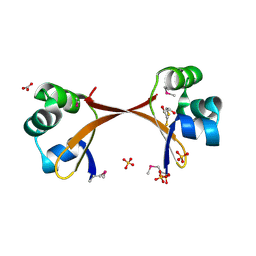 | | Structure of the Cro protein from prophage Pfl 6 in Pseudomonas fluorescens Pf-5 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BICARBONATE ION, Prophage Pfl 6 Cro, ... | | Authors: | Roessler, C.G, Roberts, S.A, Montfort, W.R, Cordes, M.H.J. | | Deposit date: | 2007-04-13 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3AYX
 
 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an H2-reduced condition | | Descriptor: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
6PQP
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist BITC | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-benzylthioformamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
3W1R
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-045a | | Descriptor: | 2,6-dioxo-5-(2-phenylethyl)-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-20 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-045a
To be Published
|
|
3W22
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-125 | | Descriptor: | 5-[2-(4-tert-butylphenyl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-125
To be Published
|
|
