2GH5
 
 | | Crystal Structure of human Glutathione Reductase complexed with a Fluoro-Analogue of the Menadione Derivative M5 | | Descriptor: | 6-(3-METHYL-1,4-DIOXO-1,4-DIHYDRONAPHTHALEN-2-YL)HEXANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Fritz-Wolf, K, Winzer, A, Bauer, H, Schirmer, H, Davioud-Charvet, E. | | Deposit date: | 2006-03-25 | | Release date: | 2006-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fluoro analogue of the menadione derivative 6-[2'-(3'-methyl)-1',4'-naphthoquinolyl]hexanoic acid is a suicide substrate of glutathione reductase. Crystal structure of the alkylated human enzyme
J.Am.Chem.Soc., 128, 2006
|
|
3N8Z
 
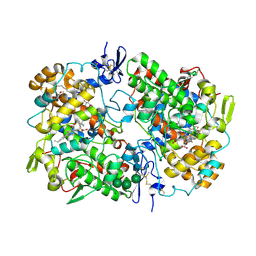 | | Crystal Structure of Cyclooxygenase-1 in Complex with Flurbiprofen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
5UTF
 
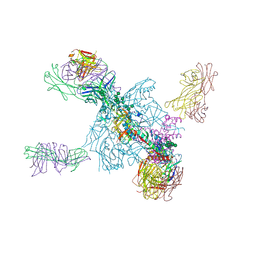 | | Crystal Structure of a Stabilized DS-SOSIP.6mut BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, Y177W, N300M, N302M, T320L, I420M in Complex with Human Antibodies PGT122 and 35O22 at 4.3 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35022 Heavy chain, ... | | Authors: | Pancera, M, Chuang, G.-Y, Xu, K, Kwong, P.D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
2GHS
 
 | |
2JE4
 
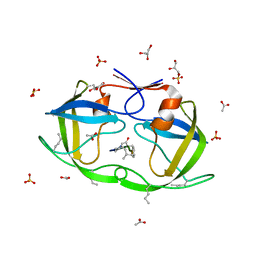 | | Atomic-resolution crystal structure of chemically-synthesized HIV-1 protease in complex with JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Modular Total Chemical Synthesis of a Human Immunodeficiency Virus Type 1 Protease.
J.Am.Chem.Soc., 129, 2007
|
|
2PFG
 
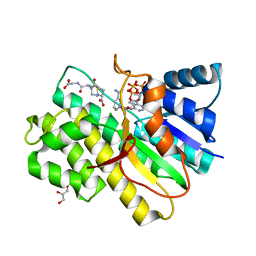 | | Crystal structure of human CBR1 in complex with BiGF2. | | Descriptor: | (5R,10S)-5-{[(CARBOXYMETHYL)AMINO]CARBONYL}-7-OXO-3-THIA-1,6-DIAZABICYCLO[4.4.1]UNDECANE-10-CARBOXYLIC ACID, CHLORIDE ION, Carbonyl reductase [NADPH] 1, ... | | Authors: | Rauh, D, Bateman, R.L, Shokat, K.M. | | Deposit date: | 2007-04-04 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Glutathione traps formaldehyde by formation of a bicyclo[4.4.1]undecane adduct.
Org.Biomol.Chem., 5, 2007
|
|
1GPE
 
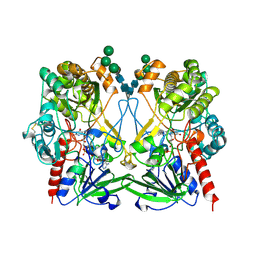 | | GLUCOSE OXIDASE FROM PENICILLIUM AMAGASAKIENSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hendle, J, Kalisz, H.M, Hecht, H.J. | | Deposit date: | 1999-03-24 | | Release date: | 1999-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2F92
 
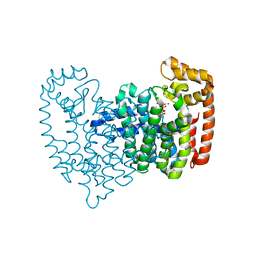 | | Crystal structure of human FPPS in complex with alendronate | | Descriptor: | 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, Farnesyl Diphosphate Synthase, PHOSPHATE ION, ... | | Authors: | Rondeau, J.-M, Bitsch, F, Bourgier, E, Geiser, M, Hemmig, R, Kroemer, M, Lehmann, S, Ramage, P, Rieffel, S, Strauss, A, Green, J.R, Jahnke, W. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs.
Chemmedchem, 1, 2006
|
|
1ARP
 
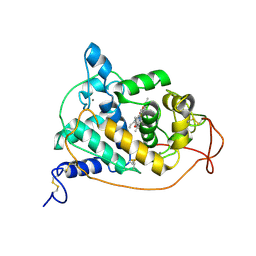 | |
3RKE
 
 | | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution | | Descriptor: | (4-aminophenyl)-imidazol-1-yl-methanone, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dube, D, Singh, R.P, Sinha, M, Singh, A.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution
TO BE PUBLISHED
|
|
1B8G
 
 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE | | Descriptor: | PROTEIN (1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Capitani, G, Hohenester, E, Feng, L, Storici, P, Kirsch, J.F, Jansonius, J.N. | | Deposit date: | 1999-01-31 | | Release date: | 2000-01-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene.
J.Mol.Biol., 294, 1999
|
|
1U3W
 
 | |
1IGX
 
 | | Crystal Structure of Eicosapentanoic Acid Bound in the Cyclooxygenase Channel of Prostaglandin Endoperoxide H Synthase-1. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,8,11,14,17-EICOSAPENTAENOIC ACID, ... | | Authors: | Malkowski, M.G, Thuresson, E.D, Smith, W.L, Garavito, R.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of eicosapentaenoic and linoleic acids in the cyclooxygenase site of prostaglandin endoperoxide H synthase-1.
J.Biol.Chem., 276, 2001
|
|
3R55
 
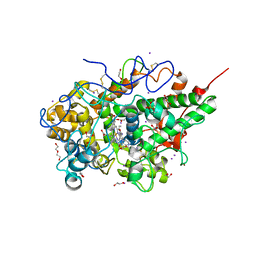 | | Crystal structure of the complex of goat lactoperoxidase with Pyrazinamide at 2.1 A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, R.P, Pandey, N, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-03-18 | | Release date: | 2011-08-03 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with Pyrazinamide at 2.1 A resolution
To be Published
|
|
1IGZ
 
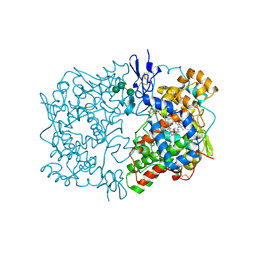 | | Crystal Structure of Linoleic acid Bound in the Cyclooxygenase Channel of Prostaglandin Endoperoxide H Synthase-1. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Malkowski, M.G, Thuresson, E.D, Smith, W.L, Garavito, R.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of eicosapentaenoic and linoleic acids in the cyclooxygenase site of prostaglandin endoperoxide H synthase-1.
J.Biol.Chem., 276, 2001
|
|
2EAD
 
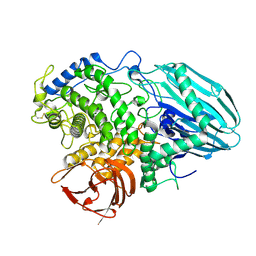 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-fucosidase, CALCIUM ION, ... | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
1U67
 
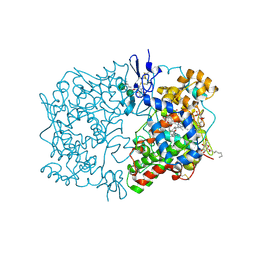 | | Crystal Structure of Arachidonic Acid Bound to a Mutant of Prostagladin H Synthase-1 that Forms Predominantly 11-HPETE. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Harman, C.A, Rieke, C.J, Garavito, R.M, Smith, W.L. | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of arachidonic Acid bound to a mutant of prostaglandin endoperoxide h synthase-1 that forms predominantly 11-hydroperoxyeicosatetraenoic Acid.
J.Biol.Chem., 279, 2004
|
|
2J9J
 
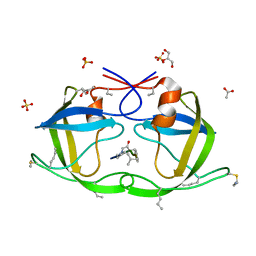 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2006-11-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
1CQE
 
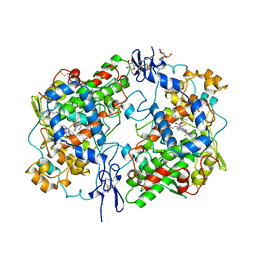 | | PROSTAGLANDIN H2 SYNTHASE-1 COMPLEX WITH FLURBIPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, PROTEIN (PROSTAGLANDIN H2 SYNTHASE-1), ... | | Authors: | Picot, D, Loll, P.J, Mulichak, A.M, Garavito, R.M. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1.
Nature, 367, 1994
|
|
1CF3
 
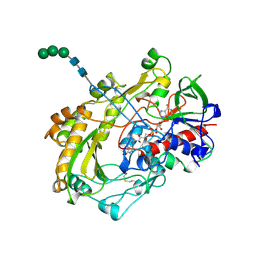 | | GLUCOSE OXIDASE FROM APERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (GLUCOSE OXIDASE), ... | | Authors: | Hecht, H.J, Kalisz, H. | | Deposit date: | 1999-03-23 | | Release date: | 1999-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1AQ0
 
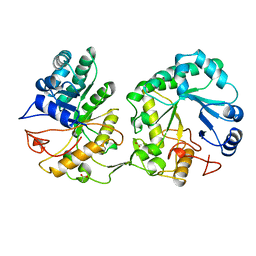 | | BARLEY 1,3-1,4-BETA-GLUCANASE IN MONOCLINIC SPACE GROUP | | Descriptor: | 1,3-1,4-BETA-GLUCANASE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION | | Authors: | Mueller, J.J, Thomsen, K.K, Heinemann, U. | | Deposit date: | 1997-08-05 | | Release date: | 1998-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of barley 1,3-1,4-beta-glucanase at 2.0-A resolution and comparison with Bacillus 1,3-1,4-beta-glucanase.
J.Biol.Chem., 273, 1998
|
|
1EPA
 
 | |
1ELA
 
 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-prolinamide, ACETIC ACID, CALCIUM ION, ... | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-04-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
4CFS
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH ITS NATURAL SUBSTRATE 1-H-3-HYDROXY-4- OXOQUINALDINE | | Descriptor: | 1-H-3-HYDROXY-4-OXOQUINALDINE 2,4-DIOXYGENASE, 3-HYDROXY-2-METHYLQUINOLIN-4(1H)-ONE, D(-)-TARTARIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Origin of the Proton-Transfer Step in the Cofactor-Free 1-H-3-Hydroxy-4-Oxoquinaldine 2,4- Dioxygenase: Effect of the Basicity of an Active Site His Residue.
J.Biol.Chem., 289, 2014
|
|
4CGL
 
 | | Leishmania major N-myristoyltransferase in complex with an aminoacylpyrrolidine inhibitor | | Descriptor: | (3R)-3-azanyl-4-(4-chlorophenyl)-1-[(3S,4R)-3-(4-chlorophenyl)-4-(hydroxymethyl)pyrrolidin-1-yl]butan-1-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
