1TMO
 
 | | TRIMETHYLAMINE N-OXIDE REDUCTASE FROM SHEWANELLA MASSILIA | | Descriptor: | GUANYLATE-O'-PHOSPHORIC ACID MONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,5,6,7,8A,9,10,10A-OCTAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL) ESTER, MOLYBDENUM (IV)OXIDE, TRIMETHYLAMINE N-OXIDE REDUCTASE | | Authors: | Czjzek, M, Dos Santos, J.P, Giordano, G, Mejean, V. | | Deposit date: | 1998-08-03 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of oxidized trimethylamine N-oxide reductase from Shewanella massilia at 2.5 A resolution.
J.Mol.Biol., 284, 1998
|
|
9DHG
 
 | | DHODH in complex with Compound 5 | | Descriptor: | (3P,7P)-7-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-6-fluoro-3-(2-methylphenyl)-1-(propan-2-yl)quinolin-4(1H)-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Identification of isoquinolinone DHODH inhibitor isosteres.
Bioorg.Med.Chem.Lett., 113, 2024
|
|
4P7X
 
 | | L-pipecolic acid-bound L-proline cis-4-hydroxylase | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-OXOGLUTARIC ACID, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ... | | Authors: | Shomura, Y, Koketsu, K, Moriwaki, K, Hayashi, M, Mitsuhashi, S, Hara, R, Kino, K, Higuchi, Y. | | Deposit date: | 2014-03-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Refined Regio- and Stereoselective Hydroxylation of l-Pipecolic Acid by Protein Engineering of l-Proline cis-4-Hydroxylase Based on the X-ray Crystal Structure.
Acs Synth Biol, 4, 2015
|
|
7AQ3
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583D | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
6M35
 
 | | Crystal structure of sulfur oxygenase reductase from Sulfurisphaera tokodaii | | Descriptor: | FE (III) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sato, Y, Yabuki, T, Arakawa, T, Yamada, C, Fushinobu, S, Wakagi, T. | | Deposit date: | 2020-03-02 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase fromSulfurisphaera tokodaii.
J Struct Biol X, 4, 2020
|
|
7AQ4
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583E | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
4NSE
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, L-ARG COMPLEX | | Descriptor: | ACETATE ION, ARGININE, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1998-10-07 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center.
Cell(Cambridge,Mass.), 95, 1998
|
|
7APY
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, D576A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQA
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H382A | | Descriptor: | (dicuprio-$l^{3}-sulfanyl)-sulfanyl-copper, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
4Y9J
 
 | |
4YLW
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wu, D, Ouyang, P, Lu, W, Huang, J. | | Deposit date: | 2015-03-06 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound
To Be Published
|
|
6LSV
 
 | | Crystal structure of JOX2 in complex with 2OG, Fe, and JA | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Probable 2-oxoglutarate-dependent dioxygenase At5g05600, ... | | Authors: | Zhang, X, Wang, D, Liu, J. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure-guided analysis of Arabidopsis JASMONATE-INDUCED OXYGENASE (JOX) 2 reveals key residues for recognition of jasmonic acid substrate by plant JOXs.
Mol Plant, 14, 2021
|
|
7JK7
 
 | | Crystal structure of Importin alpha 2 in complex with LSD1 NLS S111E mutant | | Descriptor: | GLYCEROL, Importin subunit alpha-1, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McCuaig, R, Tan, A.H.Y, Hardy, K, Seddiki, N, Ali, S, Dahlstron, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K.M, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
9EG9
 
 | | Crystal structure of human dihydroorotate dehydrogenase in complex with lapachol | | Descriptor: | 2-hydroxy-3-(3-methylbut-2-en-1-yl)naphthalene-1,4-dione, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Purificacao, A.D, Benz, L.S, Weiss, M.S, Nonato, M.C. | | Deposit date: | 2024-11-21 | | Release date: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic Structure of Human Dihydroorotate Dehydrogenase in Complex with the Natural Product Inhibitor Lapachol.
Acs Omega, 10, 2025
|
|
5BKD
 
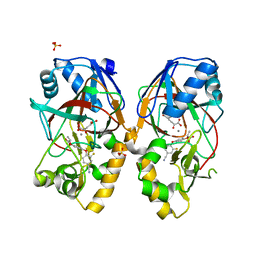 | | Crystal structure of AAD-1 in complex with (R)-cyhalofop, Mn(II), and 2-oxoglutarate | | Descriptor: | (2R)-2-[4-(4-cyano-2-fluorophenoxy)phenoxy]propanoic acid, (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, 2-OXOGLUTARIC ACID, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2019-06-02 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6BVQ
 
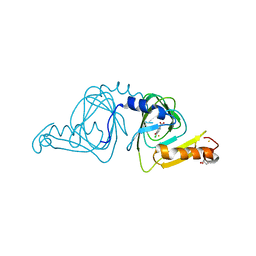 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase N27A from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2017-12-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
3MDT
 
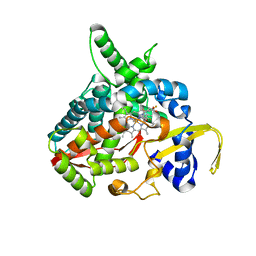 | | Voriconazole complex of Cytochrome P450 46A1 | | Descriptor: | Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
7C3L
 
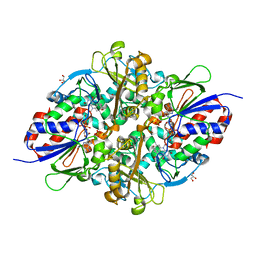 | | Structure of L-lysine oxidase D212A/D315A in complex with L-tyrosine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-lysine oxidase, ... | | Authors: | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
7T5Y
 
 | | E. coli dihydroorotate dehydrogenase bound to the inhibitor HMNQ | | Descriptor: | 3-methyl-2-[(2E)-non-2-en-1-yl]quinolin-4(1H)-one, Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Horwitz, S.M, Ambarian, J.A, Davis, K.M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines.
Rsc Chem Biol, 3, 2022
|
|
7AQ6
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583F | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ5
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583N | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ7
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583Y | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
5FNO
 
 | | Manganese Lipoxygenase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, ... | | Authors: | Wennman, A, Karkehabadi, S, Oliw, E.H, Chen, Y. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-27 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.038 Å) | | Cite: | Crystal Structure of Manganese Lipoxygenase of the Rice Blast Fungus Magnaporthe Oryzae
J.Biol.Chem., 291, 2016
|
|
9ENH
 
 | | L-amino acid oxidase 4 (HcLAAO4) from the fungus Hebeloma cylindrosporum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase 4, S-1,2-PROPANEDIOL, ... | | Authors: | Gilzer, D, Koopmeiners, S, Fischer von Mollard, G, Niemann, H.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and enzyme engineering of the broad substrate spectrum l-amino acid oxidase 4 from the fungus Hebeloma cylindrosporum.
Febs Lett., 598, 2024
|
|
6FO6
 
 | | CryoEM structure of bovine cytochrome bc1 in complex with the anti-malarial inhibitor SCR0911 | | Descriptor: | 7-methoxy-3-methyl-2-[5-[4-(trifluoromethyloxy)phenyl]pyridin-3-yl]quinolin-4-ol, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-06 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
