2E9Q
 
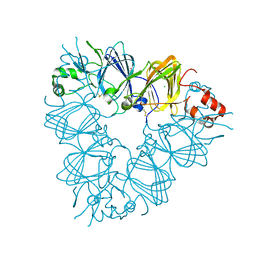 | | Recombinant pro-11S globulin of pumpkin | | Descriptor: | 11S globulin subunit beta, CHLORIDE ION, PHOSPHATE ION | | Authors: | Fukuda, T, Prak, K, Itoh, T, Masuda, T, Maruyama, N, Mikami, B, Utsumi, S. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
2E9R
 
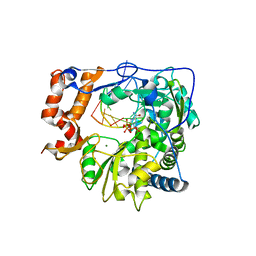 | | Foot-and-mouth disease virus RNA-dependent RNA polymerase in complex with a template-primer RNA and with ribavirin | | Descriptor: | 5'-R(*CP*AP*UP*GP*GP*GP*CP*CP*C)-3', 5'-R(*CP*CP*C*GP*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E9S
 
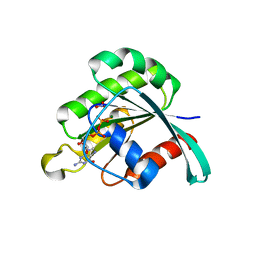 | | human neuronal Rab6B in three intermediate forms | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Vellieux, F.M, Tcherniuk, S, Garcia-Saez, I, Kozielski, F. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 3D structure of human neuronal Rab6B in three intermediate forms
To be Published
|
|
2E9T
 
 | | Foot-and-mouth disease virus RNA-polymerase RNA dependent in complex with a template-primer RNA and 5F-UTP | | Descriptor: | 5'-R(*GP*GP*GP*CP*CP*CP*(5FU))-3', 5'-R(P*UP*AP*GP*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E9U
 
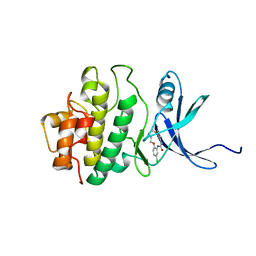 | | Structure of h-CHK1 complexed with A780125 | | Descriptor: | 18-CHLORO-11,12,13,14-TETRAHYDRO-1H,10H-8,4-(AZENO)-9,15,1,3,6-BENZODIOXATRIAZACYCLOHEPTADECIN-2-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-27 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis and Biological Evaluation of Potent Selective Macrocyclic Chk1 Inhibitors
To be Published
|
|
2E9V
 
 | | Structure of h-CHK1 complexed with A859017 | | Descriptor: | 18-CHLORO-2-OXO-17-[(PYRIDIN-4-YLMETHYL)AMINO]-2,3,11,12,13,14-HEXAHYDRO-1H,10H-4,8-(AZENO)-9,15,1,3,6-BENZODIOXATRIAZACYCLOHEPTADECINE-7-CARBONITRILE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-27 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis and Biological Evaluation of Potent Selective Macrocyclic Chk1 Inhibitors
To be Published
|
|
2E9W
 
 | | Crystal structure of the extracellular domain of Kit in complex with stem cell factor (SCF) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Kit ligand, Mast/stem cell growth factor receptor | | Authors: | Yuzawa, S, Opatowsky, Y, Zhang, Z, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2007-01-27 | | Release date: | 2007-08-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis for Activation of the Receptor Tyrosine Kinase KIT by Stem Cell Factor
Cell(Cambridge,Mass.), 130, 2007
|
|
2E9X
 
 | | The crystal structure of human GINS core complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, GINS complex subunit 3, ... | | Authors: | Kamada, K, Hanaoka, F. | | Deposit date: | 2007-01-27 | | Release date: | 2007-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the human GINS complex and its assembly and functional interface in replication initiation
Nat.Struct.Mol.Biol., 14, 2007
|
|
2E9Y
 
 | |
2E9Z
 
 | | Foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA, ATP and UTP | | Descriptor: | 5'-R(*CP*AP*UP*GP*GP*GP*CP*CP*C)-3', 5'-R(*GP*GP*GP*CP*CP*CP*A)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-01-29 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EA0
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli in complex with AP-site containing DNA substrate | | Descriptor: | 5'-D(P*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(P*GP*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharov, D.O, Grollman, A.P, Shoham, G. | | Deposit date: | 2007-01-29 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Active site plasticity of endonuclease VIII: Conformational changes compensating for different substrate and mutations of critical residues
To be Published
|
|
2EA1
 
 | | Crystal structure of Ribonuclease I from Escherichia coli COMPLEXED WITH GUANYLYL-2(PRIME),5(PRIME)-GUANOSINE | | Descriptor: | GUANYLYL-2',5'-PHOSPHOGUANOSINE, Ribonuclease I | | Authors: | Zhou, K, Pan, J, Padmanabhan, S, Lim, R.W, Lim, L.W. | | Deposit date: | 2007-01-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Ribonuclease I from Escherichia Coli Complexed with Guanylyl-2(Prime),5(Prime)-Guanosine at 1.80 Angstroms Resolution
To be Published
|
|
2EA2
 
 | | h-MetAP2 complexed with A773812 | | Descriptor: | 3-ETHYL-6-{[(4-FLUOROPHENYL)SULFONYL]AMINO}-2-METHYLBENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lead optimization of methionine aminopeptidase-2 (MetAP2) inhibitors containing sulfonamides of 5,6-disubstituted anthranilic acids
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2EA3
 
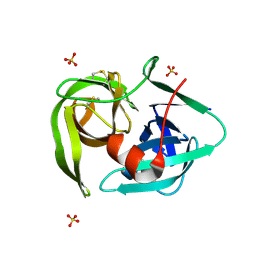 | |
2EA4
 
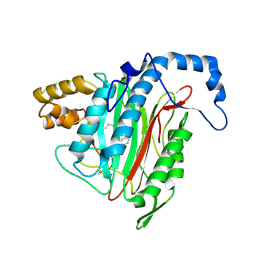 | | h-MetAP2 complexed with A797859 | | Descriptor: | 2-(2-AMINOETHOXY)-3-ETHYL-6-{[(4-FLUOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lead optimization of methionine aminopeptidase-2 (MetAP2) inhibitors containing sulfonamides of 5,6-disubstituted anthranilic acids
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2EA5
 
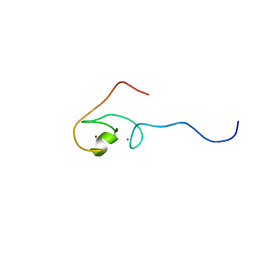 | | Solution Structure of the RING domain of the human Cell growth regulator with RING finger domain 1 protein | | Descriptor: | Cell growth regulator with RING finger domain protein 1, ZINC ION | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-30 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RING domain of the human Cell growth regulator with RING finger domain 1 protein
To be Published
|
|
2EA6
 
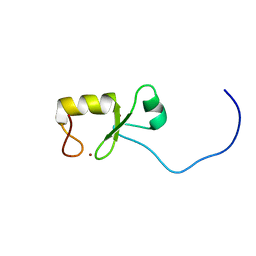 | | Solution Structure of the RING domain of the human ring finger protein 4 | | Descriptor: | RING finger protein 4, ZINC ION | | Authors: | Miyamoto, K, Yoneyama, M, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-30 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RING domain of the human ring finger protein 4
To be Published
|
|
2EA7
 
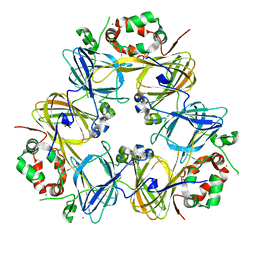 | | Crystal Structure of Adzuki Bean 7S Globulin-1 | | Descriptor: | 7S globulin-1, ACETIC ACID, CALCIUM ION | | Authors: | Fukuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and crystallography of recombinant 7S globulins of Adzuki bean and structure-function relationships with 7S globulins of various crops.
J.Agric.Food Chem., 56, 2008
|
|
2EA9
 
 | |
2EAA
 
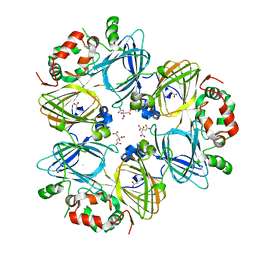 | | Crystal Structure of Adzuki Bean 7S Globulin-3 | | Descriptor: | 7S globulin-3, ACETIC ACID, CALCIUM ION, ... | | Authors: | Fukuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization and crystallography of recombinant 7S globulins of Adzuki bean and structure-function relationships with 7S globulins of various crops.
J.Agric.Food Chem., 56, 2008
|
|
2EAB
 
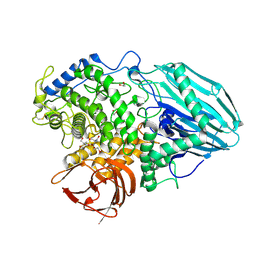 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-fucosidase, ... | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
2EAC
 
 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, Alpha-fucosidase, CALCIUM ION | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
2EAD
 
 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-fucosidase, CALCIUM ION, ... | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
2EAE
 
 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complexes with products | | Descriptor: | Alpha-fucosidase, CALCIUM ION, alpha-L-fucopyranose, ... | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
2EAK
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with lactose | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, Galectin-9, ... | | Authors: | Nagae, M, Nakamura-Tsuruta, S, Nishi, N, Nakamura, T, Hirabayashi, J, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the human galectin-9 N-terminal carbohydrate recognition domain reveals unexpected properties that differ from the mouse orthologue.
J.Mol.Biol., 375, 2008
|
|
