2ETE
 
 | | Recombinant oxalate oxidase in complex with glycolate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYOXYLIC ACID, MANGANESE (II) ION, ... | | Authors: | Opaleye, O, Rose, R.-S, Whittaker, M.M, Woo, E.-J, Whittaker, J.W, Pickersgill, R.W. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and spectroscopic studies shed light on the mechanism of oxalate oxidase
J.Biol.Chem., 281, 2006
|
|
8YHR
 
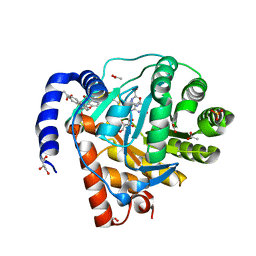 | | DHODH in complex with furocoumavirin | | Descriptor: | 4-methyl-8-[(S)-oxidanyl(phenyl)methyl]-9-phenyl-furo[2,3-h]chromen-2-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Hara, K, Okumura, H, Nakahara, M, Sato, M, Hashimoto, H, Osada, H, Watanabe, K. | | Deposit date: | 2024-02-28 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analyses of Inhibition of Human Dihydroorotate Dehydrogenase by Antiviral Furocoumavirin.
Biochemistry, 63, 2024
|
|
7VJS
 
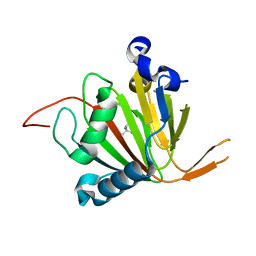 | | Human AlkB homolog ALKBH6 in complex with Tris and Ni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, NICKEL (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
9HKE
 
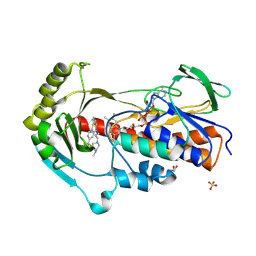 | | Crystal structure of flavin-dependent monooxygenase Tet(X4) in complex with trifluoperazine | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase, ... | | Authors: | Smith, H.G, Beech, M.J, Allen, M.D, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-12-03 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding assays enable discovery of Tet(X) inhibitors that combat tetracycline destructase resistance.
Chem Sci, 16, 2025
|
|
9HJV
 
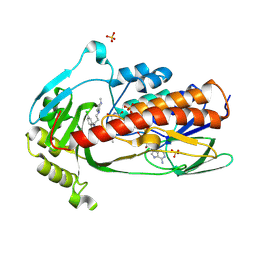 | | Crystal structure of flavin-dependent monooxygenase Tet(X4) in complex with prochlorperazine | | Descriptor: | 2-chloro-10-[3-(4-methylpiperazin-1-yl)propyl]-10H-phenothiazine, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase, ... | | Authors: | Beech, M.J, Smith, H.G, Allen, M.D, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-12-02 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding assays enable discovery of Tet(X) inhibitors that combat tetracycline destructase resistance.
Chem Sci, 16, 2025
|
|
7VXQ
 
 | | The Carbon Monoxide Complex of [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 | | Descriptor: | CARBON MONOXIDE, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Higuchi, K, Imanishi, T, Higuchi, Y. | | Deposit date: | 2021-11-13 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and spectroscopic characterization of CO inhibition of [NiFe]-hydrogenase from Citrobacter sp. S-77.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7VQS
 
 | | Crystal structure of LSD1 in complex with compound 4 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7VQT
 
 | | Crystal structure of LSD1 in complex with compound 5 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-4-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7VQU
 
 | | Crystal structure of LSD1 in complex with compound S1427 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
6J3C
 
 | | Crystal structure of human DHODH in complex with inhibitor 1291 | | Descriptor: | (6R)-1-[4-[3-(dimethylamino)phenyl]-3,5-bis(fluoranyl)phenyl]-6-propan-2-yl-6,7-dihydro-5H-benzotriazol-4-one, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | A novel series of human dihydroorotate dehydrogenase inhibitors discovered by in vitro screening: inhibition activity and crystallographic binding mode.
Febs Open Bio, 9, 2019
|
|
6J3B
 
 | | Crystal structure of human DHODH in complex with inhibitor 1289 | | Descriptor: | (6R)-1-[3,5-bis(fluoranyl)-4-[2-fluoranyl-5-(hydroxymethyl)phenyl]phenyl]-6-propan-2-yl-6,7-dihydro-5H-benzotriazol-4-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel series of human dihydroorotate dehydrogenase inhibitors discovered by in vitro screening: inhibition activity and crystallographic binding mode.
Febs Open Bio, 9, 2019
|
|
4UHA
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 3-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)-5-(methyl(2-(methylamino)ethyl)amino)benzonitrile | | Descriptor: | 3-(2-(6-AMINO-4-METHYLPYRIDIN-2-YL)ETHYL)-5-(METHYL(2-(METHYLAMINO)ETHYL)AMINO)BENZONITRILE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-03-23 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2-Aminopyridines with a Truncated Side Chain To Improve Human Neuronal Nitric Oxide Synthase Inhibitory Potency and Selectivity.
J. Med. Chem., 58, 2015
|
|
7OME
 
 | |
9VL0
 
 | | CYP105A1 complexed with simvastatin (cryogenic data collection) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Takita, T, Yoneda, S, Yasuda, K, Mizutani, K, Yasukawa, K, Sakaki, T. | | Deposit date: | 2025-06-24 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Room-temperature X-ray data collection enabled the structural determination of statin-bound CYP105A1.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7PXS
 
 | | Room temperature X-ray structure of LPMO at 1.91x10^3 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7P4H
 
 | | Crystal Structure of Monoamine Oxidase B in complex with inhibitor (+)-2 | | Descriptor: | 3,4-dimethyl-7-[[(3~{S})-piperidin-3-yl]methoxy]chromen-2-one, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Binda, C, Pisani, L. | | Deposit date: | 2021-07-11 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Reversible Coumarin Inhibitors Mutually Bound to Monoamine Oxidase B and Acetylcholinesterase Crystal Structures.
Acs Med.Chem.Lett., 13, 2022
|
|
6QQ6
 
 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) Val495Ala mutant from Alcaligenes xylosoxidans | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
7PXM
 
 | | X-ray structure of LPMO at 1.45x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PXK
 
 | | X-ray structure of LPMO at 1.39x10^5 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
8CI9
 
 | | Deoxypodophyllotoxin Synthase in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxypodophyllotoxin synthase, FE (III) ION | | Authors: | Ingold, Z, Lichman, B, Grogan, G. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure and mutation of deoxypodophyllotoxin synthase (DPS) from Podophyllum hexandrum
Front Catal, 3, 2023
|
|
8CVC
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVH
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVB
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVG
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8EVO
 
 | |
