6VGG
 
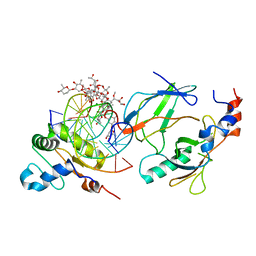 | | Crystal structure of the DNA binding domains of human transcription factor ERG, human Runx2 bound to core binding factor beta (Cbfb), and mithramycin, in complex with 16mer DNA CAGAGGATGTGGCTTC | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(P*CP*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*TP*G)-3'), ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2020-01-08 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Allosteric interference in oncogenic FLI1 and ERG transactions by mithramycins.
Structure, 29, 2021
|
|
3PX3
 
 | | Structure of TylM1 from Streptomyces fradiae H123A mutant in complex with SAH and dTDP-Quip3N | | Descriptor: | 1,2-ETHANEDIOL, N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Holden, H.M, Carney, A.E. | | Deposit date: | 2010-12-09 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Architecture of TylM1 from Streptomyces fradiae: An N,N-Dimethyltransferase Involved in the Production of dTDP-d-mycaminose .
Biochemistry, 50, 2011
|
|
3PIP
 
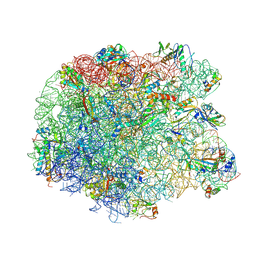 | | Crystal structure of the synergistic antibiotic pair lankamycin and lankacidin in complex with the large ribosomal subunit | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Belousoff, M.J, Shapira, T, Bashan, A, Zimmerman, E, Kinashi, H, Rozenberg, H, Yonath, A. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of the synergistic antibiotic pair, lankamycin and lankacidin, in complex with the large ribosomal subunit.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6SE7
 
 | |
3A22
 
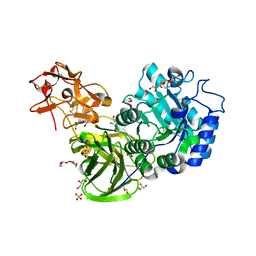 | | Crystal Structure of beta-L-Arabinopyranosidase complexed with L-arabinose | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2009-04-27 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A beta-l-Arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27.
J.Biol.Chem., 284, 2009
|
|
3A23
 
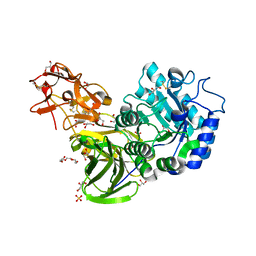 | | Crystal Structure of beta-L-Arabinopyranosidase complexed with D-galactose | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2009-04-27 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A beta-l-Arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27.
J.Biol.Chem., 284, 2009
|
|
5CFS
 
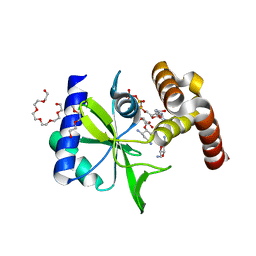 | | Crystal Structure of ANT(2")-Ia in complex with AMPCPP and tobramycin | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, AAD(2''),Gentamicin 2''-nucleotidyltransferase,Gentamicin resistance protein, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Rodionov, D, Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2015-07-08 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of the Tobramycin and Gentamicin Clinical Resistome Reveals Limitations for Next-generation Aminoglycoside Design.
Acs Chem.Biol., 11, 2016
|
|
8E18
 
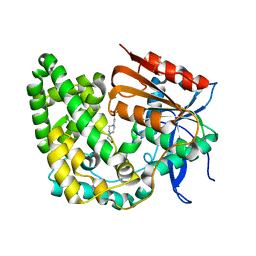 | | Crystal structure of apo TnmK1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Secreted hydrolase | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Intramolecular C-C Bond Formation Links Anthraquinone and Enediyne Scaffolds in Tiancimycin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
8E19
 
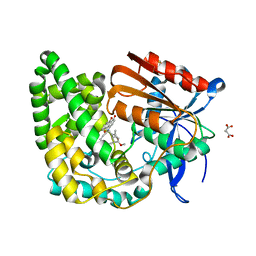 | | Crystal structure of TnmK1 complexed with TNM H | | Descriptor: | (1R,8S,13S)-8-[(4-hydroxy-9,10-dioxo-9,10-dihydroanthracen-1-yl)amino]-12-methoxy-10-methylbicyclo[7.3.1]trideca-9,11-diene-2,6-diyne-13-carbaldehyde, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SUCCINIC ACID, ... | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Intramolecular C-C Bond Formation Links Anthraquinone and Enediyne Scaffolds in Tiancimycin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
6EI0
 
 | | Cytosolic copper storage protein Csp from Streptomyces lividans: apo form | | Descriptor: | Cytosolic copper storage protein (Ccsp), GLYCEROL, SULFATE ION, ... | | Authors: | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-09-15 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
4ILV
 
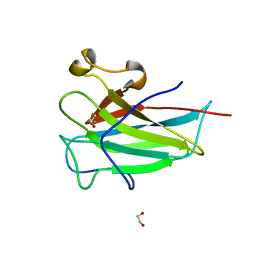 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2013-01-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
6EK9
 
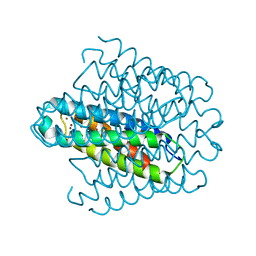 | | Cytosolic copper storage protein Csp from Streptomyces lividans: Cu loaded form | | Descriptor: | COPPER (I) ION, Cytosolic copper storage protein | | Authors: | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-09-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
5FGM
 
 | | Streptomyces coelicolor SigR region 4 | | Descriptor: | ECF RNA polymerase sigma factor SigR | | Authors: | Park, S.Y. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In Streptomyces coelicolor SigR, methionine at the -35 element interacting region 4 confers the -31'-adenine base selectivity
Biochem.Biophys.Res.Commun., 470, 2016
|
|
8E3O
 
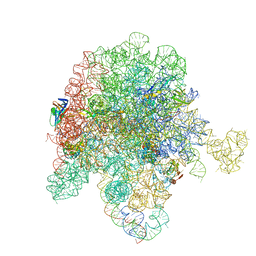 | | E. coli 50S ribosome bound to solithromycin and VM1 | | Descriptor: | (3aS,4R,7S,9R,10R,11R,13R,15R,15aR)-1-{4-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]butyl}-4-ethyl-7-fluoro-11-methoxy-3a ,7,9,11,13,15-hexamethyl-2,6,8,14-tetraoxotetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-10-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (1.99 Å) | | Cite: | Solithromycin siderophore conjugates
To Be Published
|
|
4RVH
 
 | | Crystal structure of MtmC in complex with SAH and TDP-4-keto-D-olivose | | Descriptor: | D-mycarose 3-C-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RVF
 
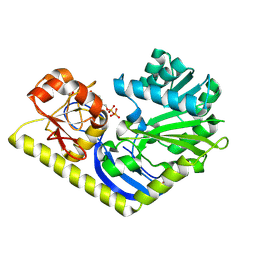 | | Crystal structure of MtmC in complex with TDP | | Descriptor: | D-mycarose 3-C-methyltransferase, THYMIDINE-5'-DIPHOSPHATE, ZINC ION | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
5DOX
 
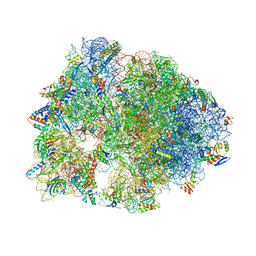 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Hygromycin-A at 3.1A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
1OS8
 
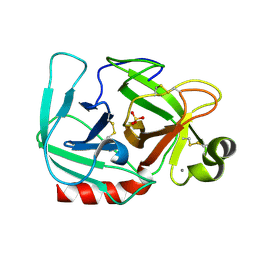 | | RECOMBINANT STREPTOMYCES GRISEUS TRYPSIN | | Descriptor: | CALCIUM ION, SULFATE ION, trypsin | | Authors: | Page, M.J, Wong, S.L, Hewitt, J, Strynadka, N.C, MacGillivray, R.T. | | Deposit date: | 2003-03-18 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Engineering the Primary Substrate Specificity of Streptomyces griseus Trypsin.
Biochemistry, 42, 2003
|
|
4ILT
 
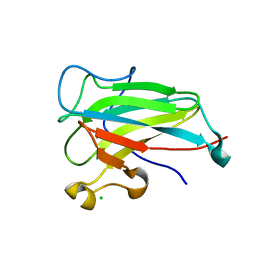 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | CHLORIDE ION, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2012-12-31 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
1SGP
 
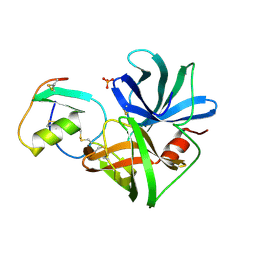 | |
5EJ3
 
 | | Crystal structure of XlnB2 | | Descriptor: | Endo-1,4-beta-xylanase B | | Authors: | Couture, J.-F. | | Deposit date: | 2015-11-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.314 Å) | | Cite: | Ligand Binding Enhances Millisecond Conformational Exchange in Xylanase B2 from Streptomyces lividans.
Biochemistry, 55, 2016
|
|
6TVO
 
 | | Human CRM1-RanGTP in complex with Leptomycin B | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shaikhqasem, A, Ficner, R. | | Deposit date: | 2020-01-10 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Characterization of Inhibition Reveals Distinctive Properties for Human andSaccharomyces cerevisiaeCRM1.
J.Med.Chem., 63, 2020
|
|
4HX6
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 | | Descriptor: | ACETATE ION, Oxidoreductase, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-09 | | Release date: | 2012-11-28 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
8E8W
 
 | |
5IT1
 
 | | Streptomyces peucetius CYP105P2 complex with biphenyl compound | | Descriptor: | 4,4'-PROPANE-2,2-DIYLDIPHENOL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2016-03-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytochrome P450 (CYP105P2) from Streptomyces peucetius and Its Conformational Changes in Response to Substrate Binding
Int J Mol Sci, 17, 2016
|
|
